Array Designer
Data Sheet
![]() Shop this product in our online store
Shop this product in our online store
Arrayit | Array Designer microarray software oligonucleotides genotyping gene expression re-sequencing CGH PCR life sciences research Arrayit and PremierBiosoft collaborate to create the most advanced oligonucleotide design software available on the market. Array Designer enables fully automated primer and probe design for VIP™ microarrays, genotyping, expression analysis, re-sequencing, CGH microarrays, real-time PCR and other applications in research, life sciences, diagnostics and agriculture. Array Designer software is fully compatible with Arrayit’s high-speed bioblue bioinformatics computers and highly recommended for VIP™ genotyping customers. Each software purchase includes Arrayit technical support for microarray applications.
Tools - Microarray Software - Array Designer Software to Design Oligonucleotides for Genotyping, Gene Expression, Re-Sequencing, CGH, and PCR Microarrays
Arrayit and PremierBiosoft collaborate to create the most advanced oligonucleotide design software available on the market. Array Designer enables fully automated primer and probe design for VIP™ microarrays, genotyping, expression analysis, re-sequencing, CGH microarrays, real-time PCR and other applications in research, life sciences, diagnostics and agriculture. Array Designer software is fully compatible with Arrayit’s high-speed bioblue bioinformatics computers and highly recommended for VIP™ genotyping customers. Each software purchase includes Arrayit technical support for microarray applications.
Table of Contents
- Introduction
- Quality Control
- Software Overview
- Software Description
- Product Contents
- Software Applications
- Software Features
- Computer Requirements
- Software Screenshots
- Customer Feedback
- Technical Assistance
- Recommended Products
- Ordering Information
- Warranty
Introduction
Arrayit and Premier Biosoft Array Designer Software improves the precision, speed and affordability of microarray experimentation in research, pharmaceutical development, diagnostics, agriculture, and other areas of life sciences and health care by facilitating both manual and automated design of primers, probes and other types of oligonucleotides. This handbook contains all the information required to take full advantage of Array Designer, the market’s most advanced oligonucleotide design software.
Quality Control
Arrayit and Premier Biosoft employ the highest quality control (QC) and quality assurance (QA) measures to ensure the quality of the algorithms used in Array Designer. The finest scientific research was used to develop this software product. When used on conjunction with Arrayit BioBlue computers, Array Designer is guaranteed to outperform the highest industry standards for oligonucleotide design software tools.
Software Overview
Array Designer is an oligonucleotide and cDNA microarray design software package that empowers scientists to design thousands of primers and probes for DNA microarray applications in seconds. It designs probes for SNP detection, microarray gene expression and gene expression profiling. In addition, comprehensive support for tiling microarrays and re-sequencing microarrays is available.
Software Description
Array Designer is the industry’s most advanced software solution for oligonucleotide design. Customers will appreciate the following features of this software package:
- Based on the most advanced microarray technology and computer science
- Complete Arrayit technical support for all microarray applications
- Manual and fully automated oligonucleotide design
- Design thousands of oligos in seconds
- Gene expression, genotyping, tiling, re-sequencing and whole genome applications
- Upload any sequence locally or globally via the Internet
- Design sequences to entire genomes effortlessly
- Optimizes sequences to avoid secondary structure
- Calculates melting temperatures and hairpin energies automatically
- Fully compatible with Arrayit microarrays and products from other vendors
- Optimized for high-speed analysis using Arrayit BioBlue Computing Platforms
- Remove the guesswork from oligonucleotide design
Product Contents
- Array Designer Software CD
- Product Registration Number
- Product Registration Sheet
Software Applications
1. BLAST Searches. Array Designer designs highly specific oligonucleotides by interpreting the results of a BLAST search performed against any genomic database available at NCBI or a local custom database. All significant homologies identified are automatically avoided during the oligonucleotide design process. Repeat regions are identified and automatically avoided as well.
2. Standard Microarray Design. You can design primers and probes for SNP detection and gene expression profiling on multiple sequences in a single search run. Our customers, typically choose over 10,000 sequences at a time. The highly specific, high-throughput capacity makes Array Designer “best in class” for oligonucleotide synthesis.
3. SNP Detection. A Single Nucleotide Polymorphism (SNP) is the variation of a single base pair in the DNA sequence between either the members of a species or between the paired chromosomes of an individual. These polymorphisms may affect how organisms develop diseases and respond to chemicals and drugs. SNPs are, therefore, of great value in biomedical research and drug development. SNP detection and genotyping can be used to explain and diagnose many diseases, to study the variation in drug responses, to establish the origin of biological material and to study the relatedness between individuals. DNA microarrays represent a technological platform that enables SNP genotyping in a rapid and massively parallel manner. SNP primers and probes for Variation Identification Platform (VIP) microarrays and other genotyping applications are optimized by Array Designer to afford optimal amplification efficiency and hybridization specificity. Array Designer designs both wild and mutant probes to amplify and genotype any allele of interest. Array Designer is highly recommended for Arrayit VIP Genotyping Platform customers. Thousands of SNPs for designing probes for SNP detection and genotyping can be loaded into Array Designer. The SNPs can be defined as a variation feature in the GenBank file format. To load unpublished SNP information, you can reformat your sequence data to a GenBank like format with the help of the template provided, or you can add SNP information to sequences already loaded in a project with the “Add SNP” option. For SNP detection assays, use the “SNP at the center of probe” option to design wild and mutant probes. Probe output can be defined to design single or multiple probes as required. For multiple probes, you can specify the sense (sense or anti-sense) and the minimum distance between the probes.
4. Gene Expression Profiling. Array Designer allows the design of thousands of specific oligonucleotides for microarray-based gene expression profiling in a single search run. Complete user control is available to alter the design parameters to meet specific research needs. You can batch BLAST sequences and avoid repeats and low complexity regions before designing sequences. The software includes automatic homology avoidance using BLAST searches of hundreds of sequences in a single search run. The results are automatically interpreted and the homologies identified are avoided during oligonucleotide design. Specificity checks include BLAST searches with primers and probes designed against all the genomic databases available at NCBI, including the redundant nr database to verify the specificity of design. Product location allows the user to specify the product length and product location at the 3’end, 5’end or anywhere in the sequence. Comprehensive selection criteria screen oligonucleotides for thermodynamic properties as well as secondary structure. Graphics for the structures are also displayed. Ranking uses statistical optimization techniques to design the best primer and probe for each template. A list of alternate designs is also available. A proprietary algorithm calculates oligonucleotide melting temperature (Tm) based on nearest-neighbor thermodynamics.
5. Whole Genome Microarrays. Whole genome microarray experiments use hybridization to scan the entire genome of an organism. The technology helps investigate expression levels of all genes, transcripts or a large fraction of the genome simultaneously with high reproducibility and sensitivity. One method of whole genome analysis uses a set of overlapping oligonucleotides covering the entire genome. The development of whole genome microaarray represents another advancement critical to the development of new microarraying applications. Whole genome microarrays have been useful for studying the poorly understood aspects of trancriptomes and natural antisense transcripts as they can simultaneously monitor gene expression on both strands of the genome. They have been found extremely useful for discovering novel genes. Array Designer allows researchers to study the genetic make-up of entire organisms effortlessly by detecting every gene or exon in the whole genome. This enables easy discovery of differentially- and alternatively-spliced transcripts, SNPs, DNA sequence variation in individuals or populations and comparative genome hybridization (CGH). Whole genome microarray design is fast and easy. can be used to interrogate a gene, exon or any other stretch of DNA of your choice. Oligonucleotides can be designed by position to detect regions of interest in the entire genome. Oligonucleotides for equal-sized fragments divides the entire genome into equal sized fragments and designs sequences for each one of them. Sequences by annotations helps you design oligonucleotides for each exon or gene. The genes and exons are resolved using GenBank file annotations or can be specified manually.
6. Tiling Microarrays. You can print every nucleotide of a long genomic sequence using amplified material obtained from whole genome amplification and avoid repetitive regions. Designing microarrays to characterize regulatory elements, or to study epigenetic modifications, methylation patterns and protein binding sites is also straightforward using Array Designer.
7. Re-sequencing Microarrays. Array Designer allows the design of re-sequencing microarrays allowing the detection of SNP and other sequence variations in a large number of samples. One of the common challenges in re-sequencing microarray design is difficult base calls caused by inter-template cross hybridization. With the unique feature called project BLAST, you can identify and avoid the homologous regions that create false positives in re-sequencing.
Software Features
Probe and primer design
- Choices in probe design: Array Designer can design single or multiple probes per sequence for your microarray projects. Allows specifying the minimum distance between two probes to avoid overlapping.
- Product Location: Allows specifying the product (amplicon) length and product location viz at the 3’end, 5’end or anywhere in the sequence.
- Comprehensive selection criteria: Screens oligonucleotides for their thermodynamic properties as well as secondary structures. Graphics for the structures are also displayed.
- Ranking: Uses statistical optimization techniques to design the best primer and probe for each template. A list of alternate designs is also available.
- Algorithm: Calculates oligonucleotide Tm using nearest-neighbor thermodynamics.
BLAST search
- Specificity Check: Thousands of searches are submitted automatically. Results are interpreted and displayed in the Homology Avoidance window.
- Batch Sequence BLAST search: Array Designer can BLAST search hundreds of sequences. The results are interpreted and the homologies identified are avoided while designing primers.
- Local BLAST: Array Designer can BLAST search against a custom local database that can be set up on a Unix machine using the WWW Standalone BLAST facility of the NCBI. Read the inbuilt product help for details.
- Desktop BLAST: BLAST search sequences against local custom databases without the need of setting up a server on a separate machine. Simply save the sequence files (.txt or .fa) on your computer and ask Array Designer to BLAST search your query sequence against them.
- Verification of specific design: The primers and probes designed by the program can be BLAST searched against all the nucleotide databases available at NCBI to verify the specificity of design.
- Repeat regions: Optimizes BLAST search parameters to detect human repeats while searching the human RNA (previously called mRNA) database at NCBI.
SNP detection and genotyping
- SNP loading: Easily add a single SNP at a time or load thousands of SNPs using standard GenBank variation files.
- SNP Detection: Designs probes for SNP detection (both wild and mutant) with SNP at the center of the probe.
- SNP genotyping with Array Designer: Designs SNP genotyping probes with SNP or one base upstream of the 3' end of the probe.
Whole genome microarrays
- By length: Probes can be designed for the fragments of a size chosen by the user. The sequence is broken down into small equal-sized fragments and then a specific probe is designed for each one of them.
- By position: Probes can be designed on genes/fragments, the start and stop position for which can be imported from a text file.
- By annotation: Probes can be designed on genes or exons, the annotations for which can be read from the GenBank file.
- BLAST to ensure specificity: For a whole genome sequence, Array Designer automatically creates a local custom database of the input sequence and BLAST searches every fragment against it. To verify the specificity of design, you can BLAST the designed oligos.
- Sequence size: Array Designer can design probes spanning the entire genome of the organism. This ability is especially useful to study the expression of intergenic and intragenic regions. Fragments to be studied can be generated on the basis of their length, their position or by specifying an annotation such as gene, exon or both.
- Uniform Tm values: Array Designer designs probes free of secondary structures and with uniform Tm values ensuring a high rate of success of microarray experiments.
Tiling microarrays
- Sequence size: Array Designer can design tiling primers to study protein binding regions of small to medium sized genomes. You can generate both distinct or overlapping amplicons depending on the specificity desired.
- Distinct amplicon: Primers are designed such that there is no overlap between adjacent amplicons.
- Overlapping amplicon: Primers are designed such that the adjacent amplicons overlap each other in the flanking regions.
- Whole Genome Amplification using Array Designer
Re-sequencing microarrays
- Resequencing probe design: In addition to standard probes, Array Designer also designs re-sequencing microarrays. With custom re-sequencing microarrays, you can detect SNP and other sequence variations in a large number of samples for applications such as biowarfare pathogen studies, , predisposition and resistance to disease or discovering the genetic basis of phenotypic traits.
- BLAST search: The challenge in designing chips for re-sequencing by hybridization is to achieve the lowest possible false positive rate. Array Designer offers an innovative and unique project BLAST capability to identify these regions and recommends a multi-chip solution.
- Avoiding repetitive regions: You can simply connect to the popular Repeat Masker using the handy link and Array Designer will avoid creating spots that may generate false positives.
Computer Requirements
Arrayit strongly recommends BioBlue Computers for Array Designer Software. The recommended computer requirements are as follows:
- Arrayit Designer version 4 and later are supported Windows 98, 2000, ME, XP, Vista, and Red Hat Linux 9.0
- Pentium IV CPU
- 512 MB of available RAM
- 1 GB of available hard disk space
- 1024 X 768 screen resolution
Software Screenshots
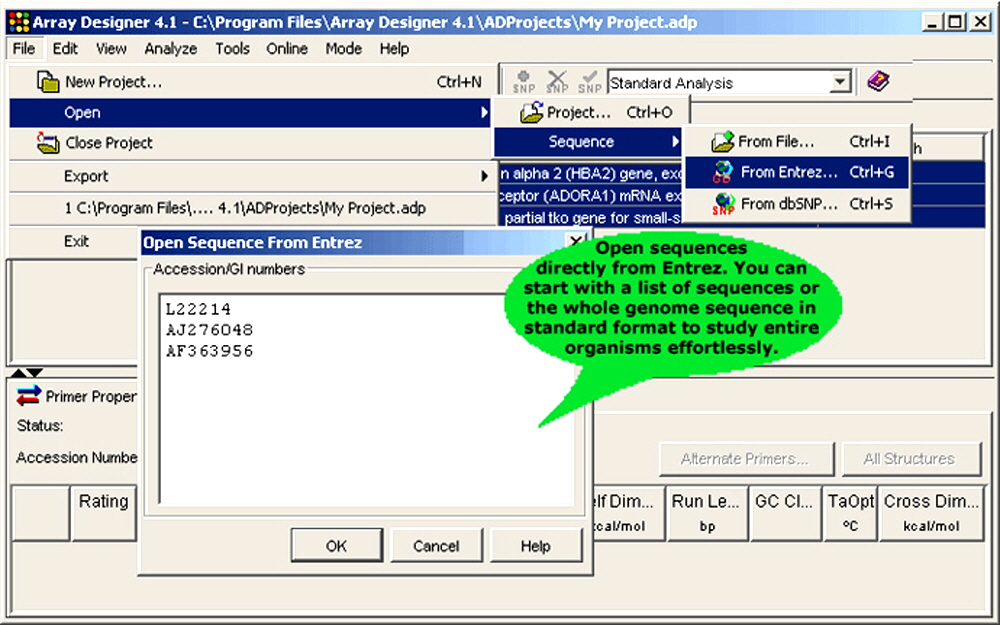
Figure 1. Array Designer is extensively integrated into the web.
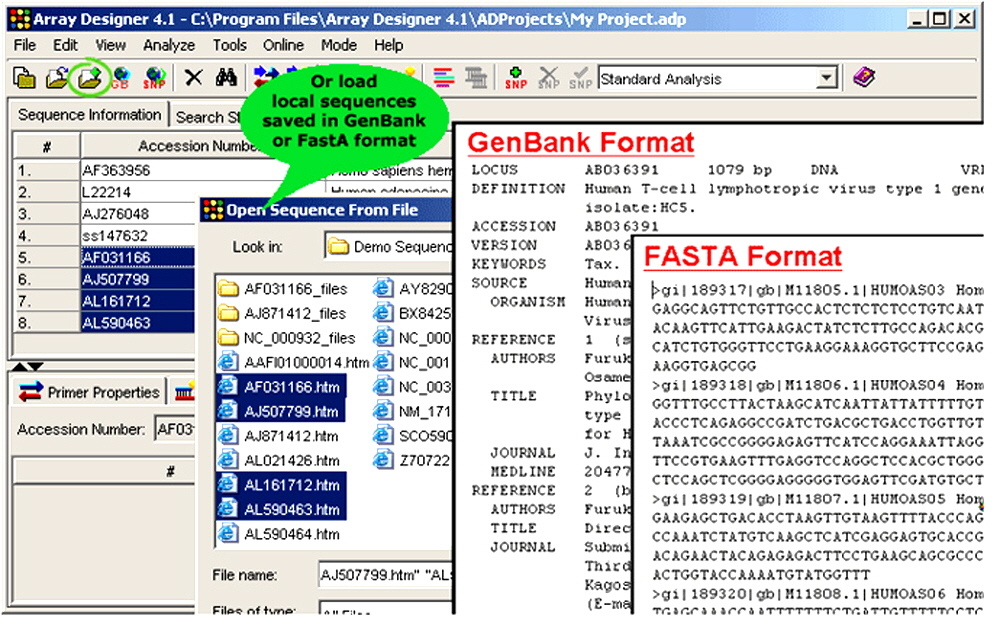
Figure 2. Array Designer also allows customers to load sequences locally.
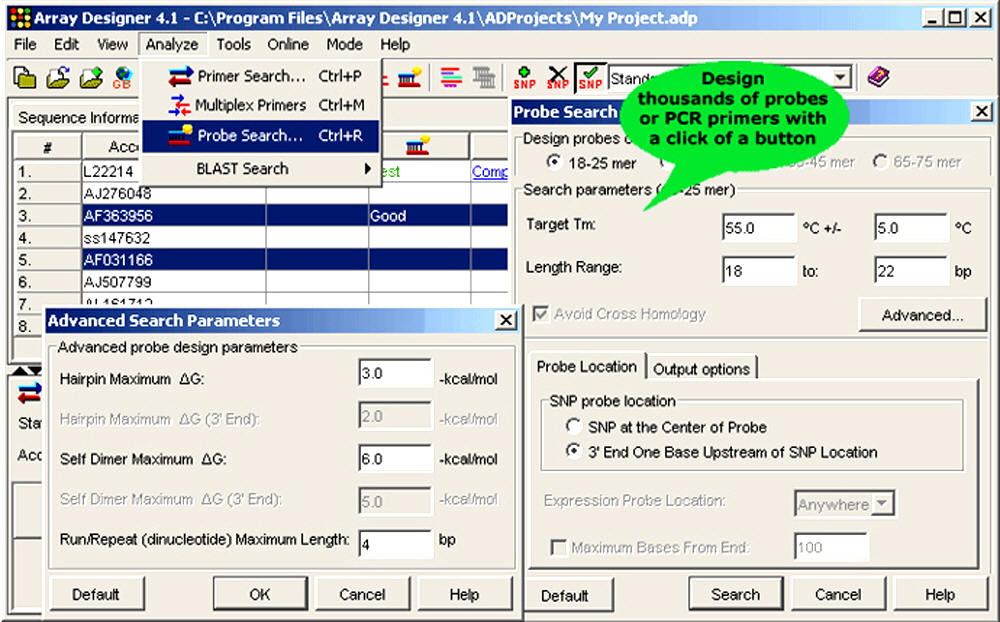
Figure 3. Array Designer also thousands of primers and probes to be designed with the click of a button.
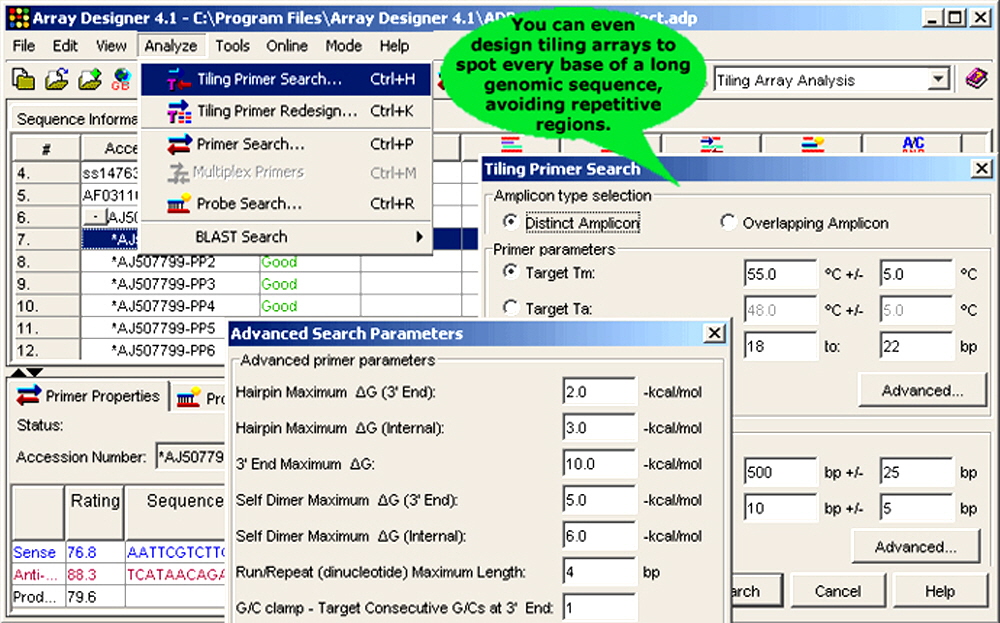
Figure 4. Array Designer has advanced features that report on melting temperature, secondary structure and other important sequence parameters for microarray applications.
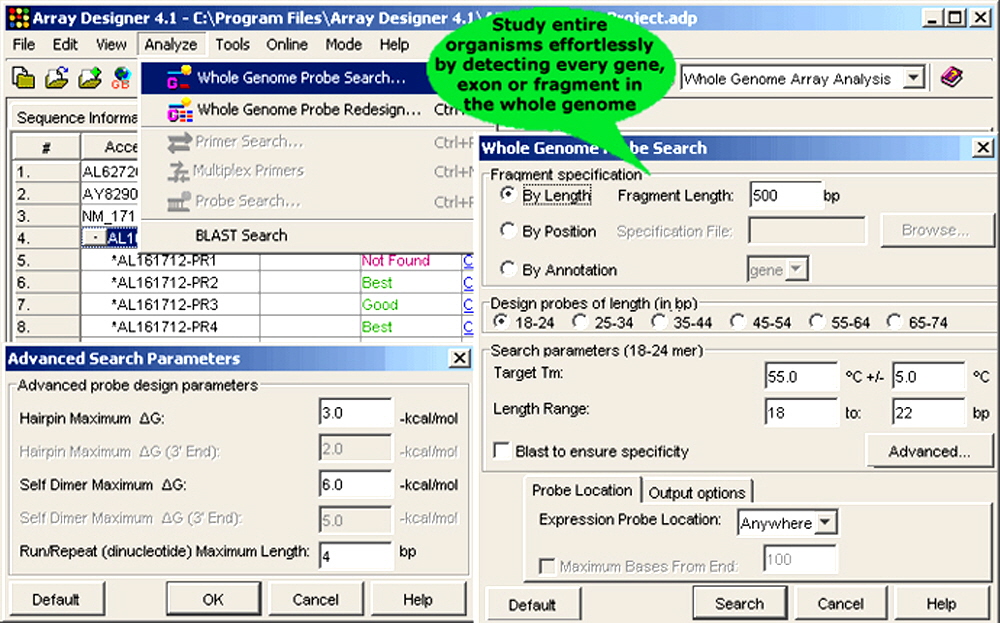
Figure 5. Array Designer empowers whole genome microarray applications.
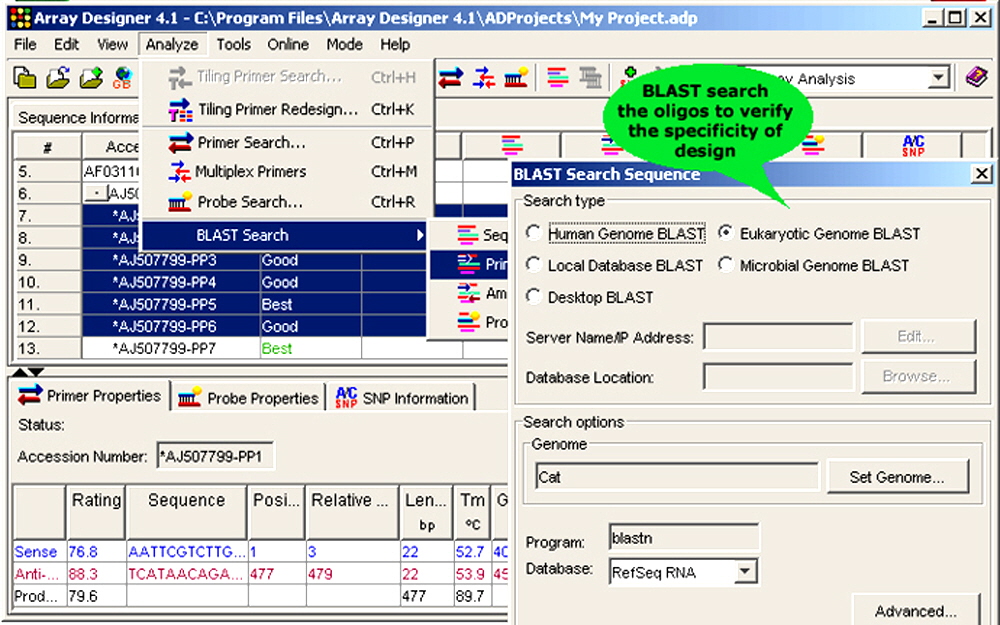
Figure 6. Array Designer has sophisticated basic local alignment search tool (BLAST) capabilities.
Customer Feedback
"With Array Designer, I was amazed at how easy it was to design primers for sequences from a cDNA library. I cloned the gene in a week. The homology avoidance algorithm works incredibly well, it made the probes so specific and my experiment a big success!"
- Dr. Greg Khitrov, Gene Array Resource Center, The Rockefeller University, New York
"The primer selection process can be reduced from weeks to days”
- HMS Beagle
"Array Designer is a very useful tool for custom designing both cDNA and oligo arrays"
- The Scientist Magazine
Technical Assistance
Please contact us if you have any questions, comments, suggestions, or if you need technical assistance. By electronic mail, use arrayit@arrayit.com and type "ArrayIt technical assistance" into the subject line. We want to hear about your successes and are always happy to feature contributed data on our website.
Recommended Products
H25K Whole Human Genome Chips
VIP Diagnostics Packages
Biomarker Discovery Services
BioBlue Bionformatics Computers
Nanoprint™ 2 Enterprise Level Microarrayers
Professional Series Microarray Printing Technology
SpotBot® 4 Personal Microarrayers
InnoScan® Microarray Laser Scanners
SpotLight™ Microarray Scanners
SpotWare™ Colorimetric Microarray Scanners

