Pathways™ Focused Human Gene Expression
Data Sheet
![]() Shop this product in our online store
Shop this product in our online store
Products - DNA Microarrays - Pathways™ Focused Human Gene Expression Microarrays for Quantitative Expression Analysis of Cultured Cells, Biopsies and LCM and DBS Specimens
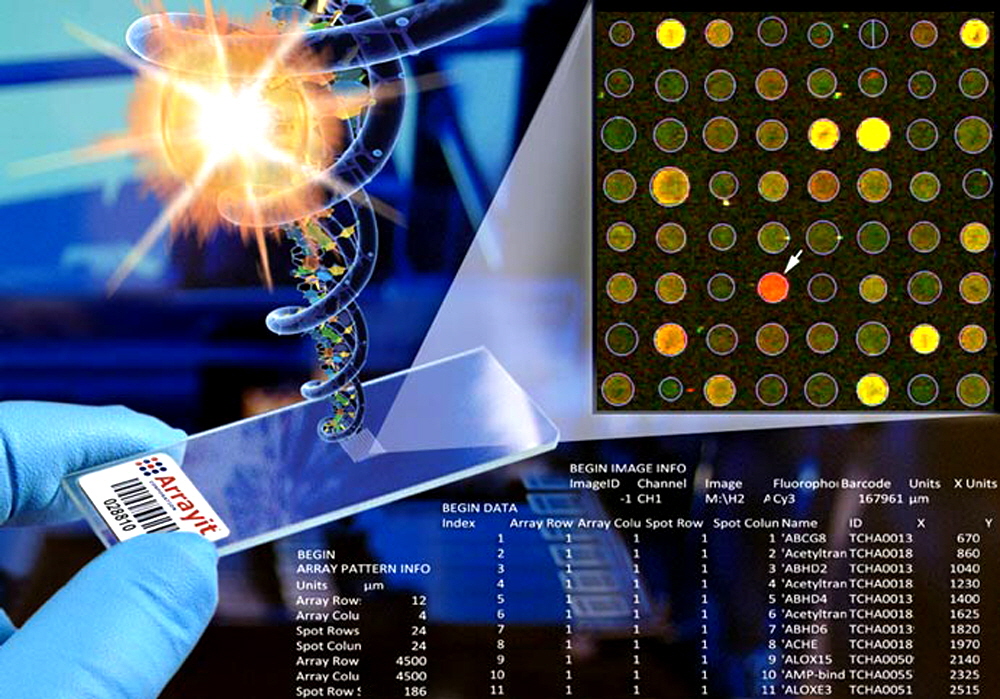
Arrayit Pathways™ Focused Human Genome Microarrays enable targeted gene expression analysis of every major cellular pathway in the human body. The content is derived from our high performance H25K human genome microarrays, which contain a complete set of 25,509 fully annotated human genes representing every gene in the human genome. Pathways™ Microarrays offer the highest sensitivity and specificity of any product on the market, empowered by genes, proprietary attachment and surface chemistry, and a unique one gene-one spot™ design. Pathways™ Microarrays allow users to rapidly and economically explore adipogenesis, angiogenesis, apoptosis, cancer drug targets, cardiotoxicity, cell cycle, cell motility, cytokine production, DNA repair, glucose metabolism, growth factors, hypertension, JAK/STAT, mitochondria, neurotoxicity, protein phoshatases, stem cells, transcription factors, WNT signalling pathways and many others. Arrayit offers a complete line of instruments, tools, kits, reagents and software to fully support Pathways™ Microarrays experimentation.
Contents
- Introduction
- Quality Control
- Product Description
- Kit Contents
- Technical Notes
- Short Protocols
- Complete Protocols
- Technical Assistance
- Troubleshooting Tips
- Recommended Equipment
- Ordering Information
- Storage Conditions
- Warranty
Introduction
The use of ArrayIt® brand products will improve the precision, speed and affordability of your microarray research in genomics, proteomics, biomedicine, pharmaceuticals and diagnostics. This handbook contains all the information required to take full advantage of Arrayit's Pathway Focused DNA Microarrays.
Quality Control
Arrayit uses the highest quality control (QC) and quality assurance (QA) measures to ensure the quality of our ArrayIt® Pathways Human DNA Microarrays. The finest science and engineering was used to develop this product. The use of advanced bioinformatics, ultra-pure oligonucleotides, superior surface chemistry and the world’s most advanced microarray cleanroom manufacturing facilities guarantees that Pathways™ Microarrays outperform the highest industry standards.
Product Description
Arrayit Pathways™ Focused Microarrays represent the most advanced human DNA microarrays in the industry. These microarrays are designed using the complete, fully annotated human genome sequence using sophisticated one spot-one gene™ informatics, useding highest gene content available. These focused microarrays are manufactured in advanced class 1 cleanrooms using our patented contact printing technology (U.S. 6,101,946), with the strictest QC and QA procedures. All our microarrays offer quality and performance that cannot be obtained from any other microarray, PCR or next generation sequencing approach. These microarrays are compatible with all slide based microarray scanners.
Users will appreciate the following features of the Pathways Focused DNA Microarrays:
- Designed from the complete, fully annotated human genome sequence
- Annotated across GenBank, UniGene, GoldenPath, RefSeq and AceView
- Made using long oligonucleotide elements
- Each gene target printed in triplicate to enhance statistical analysis
- Novel one spot-one gene™ design streamlines data analysis
- Explore the function human genes in your targeted area of interest (see list below)
- Devoid of pseudogenes, and redundant and chimeric sequences
- Control sequences add precision and reliability
- Contains widely used Ambion and Stratagene controls
- Detect many human mRNAs in a single hybridization reaction
- Manufactured using Arrayit’s patented printing technology (U.S. 6,101,946)
- Oligonucleotides made “off-line” exceed 90% purity levels
- Printed on atomically smooth glass substrates for improved precision
- Proprietary series 3 Arrayit glass offers world’s lowest intrinsic fluorescence
- Compatible with an installed base of >10,000 slide scanners
- Allow validation of Affymetrix, Agilent, ABI, GE, Qiagen PCR arrays and other vendors
- 10 µl reaction volume accelerates kinetics and minimizes sample consumption
- Format compatible with all standard data quantification and mining packages
- Compatible with dozens of DNA and RNA amplification and labeling schemes
- Utilize fluorescent, colorimetric and chemilluminscent detection
- One, two, three and four color detection possible
- Detect limit: 1 mRNA per 1,000,000
- Dynamic range: >1,000,000-fold
- Chip-to-chip CVs: ±20%
- Corner chamfer provides unambiguous spatial orientation
- Barcode assists in data tracking
- Sophisticated anti-static packaging improves usability
- Produced in our automated manufacturing facilities
- Microarrays are shipped ready for hybridization.
Kit Contents
Each Pathways kit contains the following components:
- 10 x Pathways™ Focused Human Gene Expression Microarrays
- 10 x Cover Slips (18 x 18 mm)
- 1 x Pathway Gene List
- 1 x GAL File Quantification Spot Map

Figure 2. Pathways Focused DNA Microarays use the same packaging as our H25K Human Genome Microarray. Each kit manufactured and packaged in class 1 cleanrooms and heat-sealed in anti-static packaging to ensure optimal performance and indefinate shelf life if stored clean and dry.
Gene Content
Arrayit Pathways™ Microarrays utilize subsets of the gene content present on the Arrayit H25K Human Genome Microarray, which includes 26,304 features or “spots” representing 25,509 fully annotated human genes and 795 control sequences from mouse, plant and other sources. Pathways™ Microarrays oligonucleotides contain sense strand information, and therefore nucleic acid probe mixtures reacted with these microarrays must contain anti-sense molecules to hybridize to the sense oligonucleotides present on the chip. The long oligonucleotide sequences present in each feature are annotated across the AceView, Entrez, RefSeq, UniGene, KnownID, and GenBank databases. Each oligonucleotide also includes a unique MicroarrayID, which identifies the sequence on microarary and allows customers to purchase the sequence by specifying the MicroarrayID. Gene descriptions are based on the best descriptions available from GenBank and AceView. Gene lists for Pathways™ Mcroarray are provided by electronic mail at arrayit@arrayit.com.
Short Protocol (Steps 1-7)
1. Prepare a labeled probe from a biological sample.
3. React the probe sample with a Pathways™ Microarray to allow molecular binding.
4. Wash the Pathways™ Microarray to remove unbound probe molecules.
5. Detect Pathways™ Microarray signals by scanning or imaging.
6. Quantify the Pathways™ Microarray signals.
7. Perform data analysis.
Complete Protocol (Steps 1-7)
1. Prepare a labeled probe from a biological sample. Pathways™ Microarrays contain specific sets of long oligonucleotides representing genes in a specific Pathways in the human genome. These microarrays can be used to query a single probe mixture, or multiple probes prepared using multiple fluorescent labels in multi-color DNA and RNA hybridization. For gene expression applications, each oligonucleotide designed to be complimentary to the 3’ end of a gene and is optimized to hybridize to as many transcripts as possible in cases where genes express multiple mRNAs. Each spot will detect a majority of transcripts for every gene on the microarray single reaction. There are three fundamental principles that should be used for sample preparation: (1) Sample integrity must be high. DNA, mRNA and protein molecules must be full length and free of contaminating enzymes that can produce nucleic acid and protein degradation. (2) Probe labeling must be efficient. Probes should be checked prior to reactions with to ensure that the probe mixtures are highly fluorescent. The most common cause of poor performance is poor sample/probe preparation. (3) Probe mixtures must be pure. Prior to hybridization, probe mixtures should be purified to remove non-incorporated dyes and other contaminants that can increase background noise. This step is well worth the small investment in time and money, as it will greatly increase performance. Direct and indirect fluorescent and non-fluorescent labeling techniques have been described extensively in the literature and a host of commercial labeling kits are available from many different vendors. Arrayit offers an extensive line of purification, amplification and labeling kits for DNA, mRNA and protein labeling applications. For gene expression applications, Arrayit’s Micro-Total RNA Extraction Kit, MiniAmp mRNA Amplification Kit, Indirect Amino Allyl Fluorescent Labeling Kit, and Universal Reference mRNA are highly recommended for maximum signal strength.
2. React the probe sample with microarray to allow molecular binding. The microarray is located in the center and 10 mm from the top of the glass substrate slide on the barcoded side. For additional help with side orientation, the 1.4 mm cropped corner of the glass substrate (“chamfer”) should be located in the upper right corner of the glass substrate when the barcode is facing upward. Powder free nitrile gloves should be worn at all time while handling microarrays to avoid the transfer of dust, debris, hand oils, and other contaminants that can interfere with hybridization and detection. ArrayIt® Microarray Cleanroom Gloves provide superior hand protection for microarray applications. The recommended probe sample volume for Pathways™ Microarrays is 20 µl, which will form a 60 µm sample layer on the microarray when using a 18 x 18 mm lifter type cover slip. The recommended hybridization buffer is HybIt® 2 Hybridization Solution. Arrayit provides an affordable and widely used line of Hybridization Cassettes and these work very well for low to medium throughput applications. Users may also employ the Arrayit TrayMix™ S4 Automated Hybridization Stations to allow automated reactions and washing. Probe mixtures can be generated from cloned or cellular DNA, RNA or protein, and dozens of fluorescent and non-fluorescent labeling schemes have been described in the literature. For gene expression applications with Pathways™ Microarrays use 0.5 µg of amino-allyl labeled cDNA per color (1.0 µg total of green and red probe for two-color experiments) is recommended to produce strong hybridization signals. Probe mixtures should be spun at 18,000 x g for 1 min to pellet debris prior to use. The microarray substrate slide, hybridization cassette or chamber, lifter slip and probe mixture should be pre-heated to 42°C for 5 min prior to hybridization to ensure low and uniform background. Pipette the 20 µl probe mixture directly onto the center of the microarray and quickly place the lifter slip onto the probe droplet, allowing the probe mixture to sheet evenly across the surface. Probe mixtures should be reacted for 3.5 hours at 42°C for gene expression applications, though different times and temperatures can be used for probe mixtures of different concentration and complexity. Probes derived from cloned and recombinant sources will probably produce satisfactory results within 5-10 minutes because the molecular complexity of such sources is low and Pathways™ Microarrays performance is extremely high. Probe mixtures derived from complex sources such as human genomic DNA, mRNA and protein may require more than 3.5 hours to produce strong signals. In rare cases, users may opt for up to 18 hour reaction times, though probe drying and elevated background noise can become an issue with prolonged hybridizations. After the hybridization step is complete, users should proceed quickly to the wash steps to prevent probe drying and elevated background.
3. Wash microarrays to remove unbound probe molecules. After the probe binding reaction is complete, the microarray must be washed to remove unbound material. Oligonucleotide-DNA or oligonucleotide-RNA hybrids are typically washed at high-stringency to remove labeled probe molecules that have bound weakly or non-specifically. Lesser stringency can be use in cases where DNA and mRNA probe mixtures are derived from homologous species such as primate, mouse and rat. For protein binding applications, oligonucleotide-protein complexes should be washed to remove proteins that have bound weakly or non-specifically. The oligonucleotides on Pathways™ Microarrays are bound covalently to the glass substrate, and therefore the oligonucleotides themselves are stable to elevated temperature (≤70°C), low salt, and other neutral pH (pH 6-8) treatments. Users should avoid extremes in temperature and pH to avoid damaging the oligonucleotides. Protein experiments with H25K typically make use of reactions 20-37°C and washes with phosphate-buffered saline (PBS) buffers containing dilute additions of detergent (e.g. 0.1-0.01% TritonX100). Prior to washing, the lifter slip must be removed from the microarray surface to allow efficient washing and to avoid scratching the spots on the surface. For gene expression applications, quickly remove the lifter slip by submerging microarray for 5-10 sec in 42°C in Wash Buffer A and wash as follows: 5 min at 42°C in Wash Buffer A, 5 min at room temperature in Wash Buffer B, and 1 min at room temperature in Wash Buffer C, followed by a 1 sec dunk in dH2O. Washes should be performed in High-Throughput Wash Stations with moderate mixing or in the TrayMix™ S4 Hybridization Station. Users can experiment with different wash buffers and different temperatures (20-65°C) to optimize the washing process for a particular application. Following the final wash step, microarrays should be dried quickly by centrifugation for 3 sec in an Arrayit Microarray High-Speed Centrifuge in preparation for scanning.
4. Detect signals by scanning or imaging. Pathways™ Microarrays should be inserted into a microarray scanning or imaging device to acquire the signals. The Arrayit InnoScan® 710 and 900 Fluorescence Scanners and the highly affordable Arrayit SpotLight™ Microarray Fluorescence Scanner works well for this application, though all Pathways™ Microarrays are compatible with all major brands of microarray glass microscope slide microarray scanners. Scanners and imagers should be capable of at least 10 µm resolution, the lowest recommended resolution setting. Scanning at 3-5 µm resolution produces higher quality images. The selected scan area should be large enough to capature all spots in a single image. In addition to the resolution and scan area settings, correct settings should be used for wavelength(s), gain, laser intensity, PMT, and other technical parameters required to produce high quality, data-rich images. For expression analysis applications involving complex mixtures of mRNA varying greatly in concentration, multiple scanner settings (e.g. low and high sensitivity) across two or more images can be used to capture signals spanning up to 6 orders of magnitude. Multi-color experimentation involving, for example, cyanine 3 and cyanine 5 labels, obviously require data capture at two different wavelengths (e.g. 532 and 635 nm). Images should be saved as 16-bit TIFF files for quantification and downstream analysis.
5. Quantify the signals. Arrayit MAPIX® ande ImageGene quantification software is highly recommended for quantification. Use GAL file spot map provided with the microarrays. Place grid over the file, making sure that all subgrids and spots are properly aligned. MAPIX® quantification settings should be set to allow the software to find all of the printed spots in an automated manner. The following settings work well for most applications: Max position offset (% pitch) 30, Min. diameter (% Dth) 80, Max. diameter (% Dth) 190, S/B border width (pixels) 1.6 and Background diameter (pitch) 1.6. Once the grid has been accurately aligned, the quantification process should be initiated to extract data values from H25K using the photmetric calculation command. The extracted data should be saved and then opened in Microsoft Excel or some other software program for downstream analysis.
6. Perform data analysis. Properly quantified images will yield great data. Values should be analyzed using a variety of inclusion and exclusion criteria to ensure statistical relevance of all values. For most applications, background-subtracted median signal intensities provide the most accurate reporting of spot signal intensity. Data from multiple images can be compared to identify up- and down-regulated genes in expression profiling experiments. Arrayit offers highly sophisticated Genome Pathways Builder™ software analysis for Biomarker Discovery and Validation Services customers. Genome Pathways Builder™ constructs biological Pathwayss from vast quantities of expression data, providing customers with a detailed molecular understanding of a disease process. Genome Pathways Builder™ also allows pharmaceutical clients to understand the mode of drug action as well as off-targets as a means of benchmarking pharmaceutical leads prior to and during clinical trials.
Technical Assistance
Please contact us if you have any questions, comments, suggestions, or if you need technical assistance. By electronic mail, use arrayit@arrayit.com and type "H25K technical assistance" into the subject line. We want to hear about your successes and are always happy to publish contributed H25K sample data on our website.
Figure 3. Arrayit Neurotransmitter Receptors and Regulators Pathways Microarrays (Cat. NRPM) enable targeted gene expression analysis of neurotransmitter signaling in human cells.
Troubleshooting Tips
1. Weak signals
- Degraded DNA, RNA or protein. Check sample integrity.
- Poor sample labeling efficiency. Check probe mixture.
- Wash stringency too high.
2. High background fluorescence
- Probe mixture drying out during hybridization. Keep hydrated for duration.
- Probe drying after hybridization. Transfer quickly to wash steps.
- Probe volume too low. Use ≥50.0 µl for H25K.
- Purify probe mixture to remove contaminating unincorporated fluors.
- Hybridization time too long. Use a maximum time of 6 hrs.
- Poor washing procedure. Increase time or stringency.
3. Low frequency of differentially expressed genes (gene expression)
- Wash stringency too low. Reduce salt and/or increase temperature.
- Messenger RNA samples highly similar. Check samples/stimuli.
4. Artifacts observed in two-color experiments (gene expression)
- Dye effects to blame. Swap colors and re-examine data.
Recommended Equipment
- InnoScan® 710, 910 and 1100 Series Fluorescence Laser Scanners
- SpotLight™ 2 Microarray Fluorescence Scanners
- TrayMix™ S4 Automated Hybridization Stations
- Biomarker Discovery and Validation Services
- VIP™ Genotyping Packages
- Hybridization Cassettes
- Hybridization Buffers
- Microarray High Speed Centrifuge
- High Throughput Wash Station
- Microarray Cleanroom Gloves
- Microarray Cleanroom Wipes
- Microarray Air Jet
- Micro-Total RNA Extraction Kit
- MiniAmp mRNA Amplification Kit
- Universal Reference mRNA
- CheckIt Chips

