SuperAmine Slides
Data Sheet
![]() Shop this product in our online store
Shop this product in our online store
Products - Microarray Substrates & Slides - SuperAmine, SuperAmine 2, SuperAmine 2 Low Density and SuperAmine 2 UltraLow Density for DNA Microarray Manufacturing

ArrayIt® is pleased to announce SuperAmine 2, the newest version of our original SuperAmine Microarray Slide Substrates product line. ArrayIt® substrates are the only microarray substrates in the world that have a polished atomically smooth glass surface (±20 angstroms), for the ultimate in reactive group homogeneity, low background and data precision. SuperAmine 2 Substrates are manufactured to standard “open platform” dimensions (25 x 76 mm), cleaned at the atomic level in state-of-the-art class 1 cleanrooms, and treated with our ultra-pure amine surface chemistry for superior electrostatic coupling efficiency and low background. Heat-sealing in environmental proof anti-static packaging improves shelf life and eliminates moisture and static accumulation. SuperAmine 2 Substrates have a proprietary corner chamfer for unambiguous side and end orientation, which greatly improves usability during manufacturing. SuperAmine 2 products outperform standard optical quality glass from other vendors and are the slide substrates of choice for high-density oligonucleotide and cDNA microarray manufacturing applications using contact and non-contact printing methods. SuperAmine 2 products are compatible with all major brands of microarrayers and scanners. that support the microscope slide size microarray format, the standard in the industry set by Arrayit in 1997.
Table of Contents
- Introduction
- Quality Control
- Product Description
- Technical Notes
- Short Protocol
- Complete Protocol
- Technical Assistance
- Troubleshooting Tips
- Recommended Equipment
- Ordering Information
- Storage Conditions
- Warranty
Introduction
The use of ArrayIt® brand products will improve the precision, speed and affordability of your microarray research in genomics, proteomics, biomedicine, pharmaceuticals, diagnostics and agriculture. This handbook contains all the information required to take full advantage of Arrayit's ArrayIt® brand SuperAmine 2 Microarray Substrates.
Quality Control
Arrayit uses the highest quality control (QC) and quality assurance (QA) measures to ensure the quality of our ArrayIt® brand SuperAmine 2 Microarray Substrates. The finest science and engineering was used to develop this product line. The use of ultra-pure reagents, atomic-level cleaning, class 1 cleanroom manufacturing and rigorous 6-step optical analysis of every substrate guarantees that this product line outperforms the highest industry standards.
Product Description
The ArrayIt® brand SuperAmine 2 Microarray Substrates line represents the highest quality amine-containing substrate in the microarray industry. All of our substrates are polished to atomic smoothness and manufactured in a state-of-the-art class 1 cleanrooms, with strict controls on particulate, biological and static contaminants, as well as temperature and humidity control. These pristine glass substrates with reactive amine groups offer quality and performance that cannot be obtained from any other vendor.
Users will appreciate the following:
- Polished to atomic surface smoothness <±20 angstroms over 1.0 µm2
- Superior substrate flatness of <0.036 mm over 25 x 76 mm
- Only polished glass substrate/slide in the microarray industry
- Homogenous SiO2 groups provide superior silane reactivity
- Amine density of of 2 x 10^12 groups per mm^2
- Vastly superior to unpolished optical quality glass from other vendors
- Ultimate surface for ultra-high density DNA and protein microarray manufacturing including micro-mirror, photolithography and contact printing
- Used by high-volume microarray manufacturing companies
- Used by overseas companies for diagnostics
- Topological smoothness ensures uniform hybridization layer and scanning
- Manufactured in a state-of-the-art class 1 cleanrooms
- Ultra-low intrinsic fluorescence and background noise
- Can be used as a starting point for gel and membrane coatings (e.g. polyacrylamide and nitrocellulose)
- Used by major academic core facilities
- Open platform dimensions compatible with all major brands of microarrayers and scanners
- Precise physical dimensions (25 ± 0.2 mm x 76 ± 0.3 mm x 0.940 mm ± 0.025 mm)
- Proprietary corner chamfer (1.4 mm) provides unambiguous side and end orientation to simplify printing and processing
- Finished edges enhance user safety
- Excellent refractive index, transmission and hardness specifications
- Sophisticated anti-static packaging improves usability
- Offered with or without bar-codes
- Custom laser and chrome fiducials available upon request
- Product arrives “ready to print” with no additional processing required
- High-volume 100,000 piece per month manufacturing capabilities
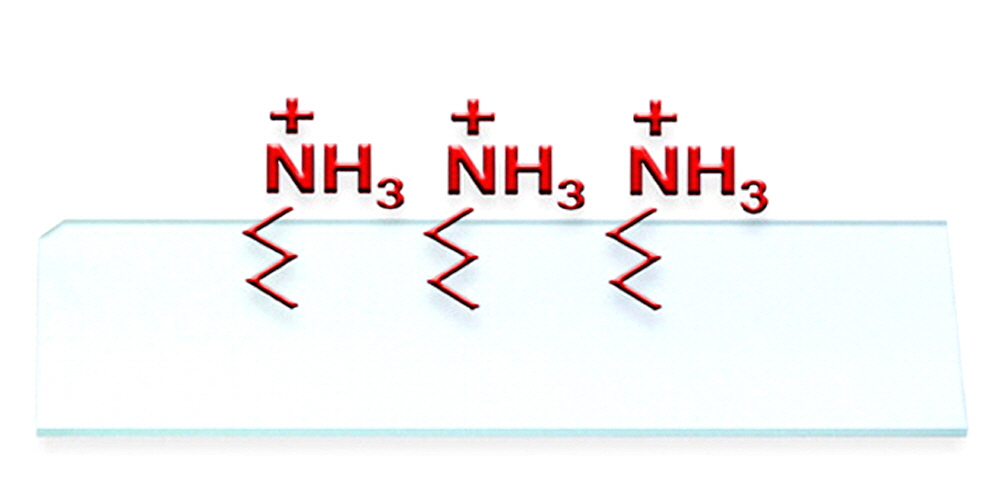
Figure 1. Schematic illustration of an ArrayIt® brand SuperAmine 2 Microarray Substrate with positively charged reactive amino groups on a polished, atomically flat glass substrate. This product is ideal for researchers and microarray manufacturing organizations engaged in oligonucleotide and cDNA microarray printing. Oligonucleotides and cDNAs bind to SuperAmine 2 via electrostatic bonds, and coupling can be enhanced by treatment with heat or ultraviolet light (see Fig. 6). The corner chamfer allows unambiguous side and end orientation, and our chemical treatment requires printing on the side shown in this diagram rather than on the backside. See Fig. 5 for additional information on the correct chamfer location relative to the printed microarray elements.
Technical Notes
ArrayIt® SuperAmine 2 Microarray Substrates are polished at high precision to achieve atomic-level smoothness of ±20 angstroms (Fig. 2). The advantages of glass polishing are several-fold. Polishing provides a uniform density of amino groups, which allows uniform coupling of the printed DNA molecules. Spots containing a uniform density of DNA molecules produce more precise signal intensities that are easily quantified. A second advantage of glass polishing is that topological smoothness reduces “peaks and valleys” in the glass, which enables uniform hybridization layer thickness on the SuperAmine 2 surface and improved precision. Unpolished surfaces inherently produce uneven hybridization layers causing signal intensities to vary due to different quantities of probe molecules available for hybridization. A third advantage of glass polishing is seen in the scanning step. Atomically flat surfaces provide a precisely calibrated distance from the glass to the scanner optics, producing highly uniform scanning data (Figs. 3 and 7). Standard optical quality glass slides and microscope slides from other vendors do not provide the advantages conferred by ArrayIt® polished glass substrates. We use the term “substrate” instead of “slide” to distinguish our products from the lesser quality microscope slides offered by other vendors.
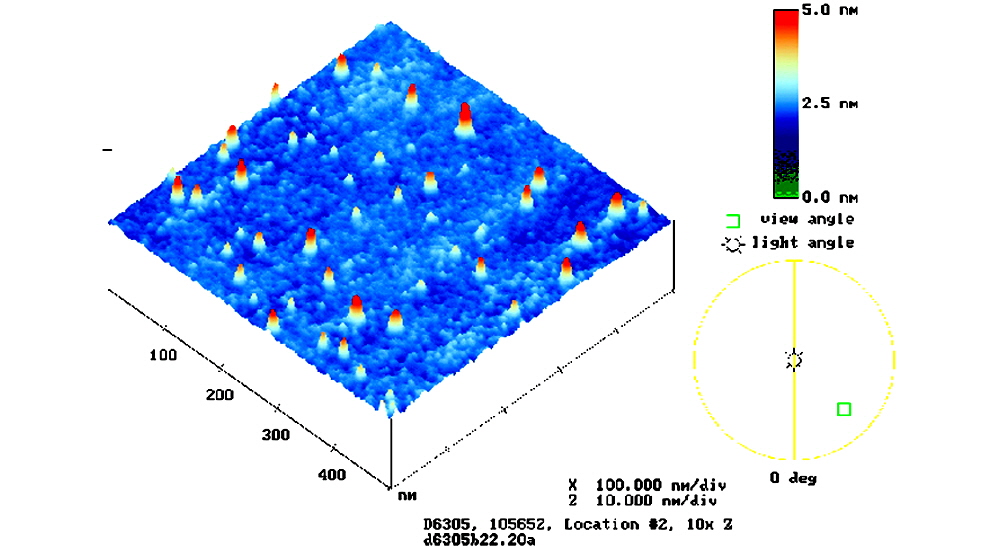
Figure 2. Shown is an atomic force microscopy (AFM) image of a SuperAmine Microarray Substrate, with smoothness data represented in a color palette. The scale bar (upper right) denotes surface smoothness in nanometers (nm). 1 nm = 10 Å. SuperAmine (shown) and SuperAmine 2 are flat to ±25 and ±20 Å, respectively.

Figure 3. Scanned image of an ArrayIt® brand SuperAmine 2 Microarray Substrate acquired at high scanner sensitivity to assess background uniformity. The SuperAmine 2 Substrate was scanned at 10 µm-resolution using a GenePix 4000A microarray scanner (Molecular Devices) set at 100% laser power and photomultiplier tube (PMT) gain 900 in the 532 nm (cyanine 3) channel. Signal intensities vary by less than ±1% over the 22 x 40 mm area. The data are represented in a color palette and the space bar corresponds to 1.0 mm.
Table 1. Shown is a technical comparison of ArrayIt® SuperAmine and SuperAmine 2 Microarray Substrates.
Specification |
SuperAmine |
SuperAmine 2 |
Atomic smoothness |
±25 Å |
±20 Å |
Glass cleaning procedure |
Dual temperature, triple phase atomic |
Triple temperature, quadruple phase atomic |
Moisture content |
Low |
Undetectable |
Optical QC procedure |
5-tier inspection and laser scanning |
6-tier inspection and laser scanning |
Intrinsic fluorescence |
Low |
Ultra-low |
Background reactivity |
Low |
Ultra-low |
Signal uniformity (22 x 72 mm area) |
±4% |
±1% |
Amino group density (per mm2) |
5 x 1012 |
2 x 1013 |
Minimum microarray feature size |
5 µm |
1 µm |
Maximum microarray feature number (22 x 72 mm area) |
15,840,000 |
396,000,000 |
Short Protocol (Steps 1-7)
1. Decontaminate shipping envelope to remove particles.
2. Transfer glass substrates onto microarrayer platen.
3. Print oligonucleotide and cDNA microarrays using contact or non-contact printing.
4. Process the printed microarrays to remove unbound molecules.
5. Hybridized labeled samples prepared from DNA, mRNA and protein sources.
6. Scan the microarrays to acquire data files.
7. Perform data analysis.
Complete Protocol (Steps 1-7)
1. Decontaminate shipping envelope to remove particles. Using ArrayIt® brand Microarray Cleanroom Wipes and the Microarray Air Jet, wipe down the silver anti-static package with 95% ethanol and blow dry to remove dust and other contaminants that may have accumulated during shipping. This cleaning process prevents contamination of the SuperAmine 2 Substrates. For cleanroom users, the substrates may be transported into the cleanroom following the topical decontamination step. Once inside the cleanroom or prior to printing, open the anti-static package by cutting between the heat seal and the zip-lock approximately 1.0 cm below the top end of the package. This will allow the silver package to be re-sealed if only a portion of the 25 substrates is used in a single print run. To open the white substrates box, lift the top gently upward while holding the bottom firmly in the other hand. Once the substrates box is opened, remove the white cleanroom cloth by lifting it gently upward. Avoid dragging the cleanroom cloth across the substrates as this may cause fraying of the cloth. All 25 substrates may be used in a single printing or a portion of the substrates may be saved for future print runs by closing the substrates box lid, placing the box inside the silver anti-static package, and re-sealing the zip-lock. SuperAmine and SuperAmine 2 Substrates are good for 6 months from the date of receipt.
2. Transfer glass substrates onto microarrayer platen. Remove each substrate from the white box and carefully transfer each onto the printing platen of the microarrayer. The corner chamfer should be use to ensure that each substrate is positioned correctly on the platen. While holding the substrate in the vertical position, the corner chamfer should occupy the upper right corner. Do not attempt to print on the backside of a SuperAmine or SuperAmine 2 Substrate. Figure 5 shows the correct placement of printed microarrays relative to the corner chamfer. Make sure each substrate rests firmly against the printing platen. The platen should be free of dust, debris, hand oils and all other contaminants that can interfere with substrate locating and printing. This is particularly important for microarray systems such as NanoPrint and SpotBot 2, in which the substrates are held in place using a spring-loaded mechanism using direct contact between multiple adjacent substrates. The failure to seat the substrates uniformly and completely against the printing platen can cause locator errors and robot crashes. One effective way to ensure proper seating is to lower your head and align your sight line (eyes) horizontally across the platen. This will allow you to make sure that each substrate occupies the same horizontal plane. By looking carefully across the horizontal printing plane, substrates that are misaligned or “buckled” are readily identified and can be corrected prior to printing.
3. Print oligonucleotide and cDNA microarrays using contact or non-contact printing. SuperAmine and SuperAmine 2 Substrates provide the ultimate starting points for microarray printing owing to the extraordinary quality of these glass surfaces. Arrayit’s patented contact printing technology, installed in >3,000 laboratories, produces microarrays of superb quality. ArrayIt® brand Nanoprint and SpotBot microarrayers represent the ultimate enterprise-level and personal instruments, respectively, for microarray manufacture. Prior to printing, samples should be free of contaminants such as salts and primers that can interfere with coupling. For cDNA microarray applications, ArrayIt® brand PCR Purification Kits allow highly affordable purification of amplified cDNAs and should be used instead of ethanol precipitation. For oligonucleotide microarrays, the oligonucleotides should be free of ammonium hydroxide and reverse phase cartridge purification works very well for most purification applications. Purified cDNAs and oligonucleotides should be re-suspended in Micro Spotting Solution Plus or another suitable printing buffer. Users should select pin sizes, pin numbers and center-to-center spacing to create microarrays of the desired spot diameter and physical size. Printed spots can be inspected by light microscopy (20X) to assess printing efficiency and spot geometry. Microarrayers equipped with imaging systems allow microarrays to be visualized in “real time” during printing.
4. Process the printed microarrays to remove unbound molecules. After printing, spots containing cDNA and oligonucleotide molecules dehydrate, and the loss of H2O results in a large increase in DNA concentration. Highly concentrated DNA on the SuperAmine and SuperAmine 2 surfaces results in electrostatic coupling between negatively charged phosphate groups on the DNA backbone and positively charged amino groups on the substrate surface (Fig. 6). Attachment to the surface can be strengthened by baking the printed substrates for 80 minutes at 80°C in a drying oven without vacuum or by cross-linking with ultraviolet light. Baking and cross-linking can be used together in cases where attachment is deemed difficult, but this combination is not recommended for cDNAs and oligonucleotides because excessive attachment can sterically interfere with hybridization and reduce signals. After attachment, the microarrays should be processed to remove unbound DNA. Rapid mixing for 60 min at room temperature (20-25°C) in a High Throughput Wash Station containing ArrayIt® BlockIt Blocking Buffer provides a very effective means of simultaneously processing and blocking the microarray surface. The advantage conferred by BlockIt is that DNA molecules removed during processing are “blocked” from re-attaching to the surface, preventing the formation of “comet tails” and providing extremely low background and very sharp and clean microarray features. After the blocking step, microarrays should be washed for an additional 30 sec in distilled H2O and spun dry using a Microarray High Speed Centrifuge or for 1 min (500 x g) in a traditional centrifuge (e.g Savant). For cDNA microarrays, denature the double-stranded DNA on the surface by immersing the substrates in distilled H2O (100°C) for 1 min, and plunge the substrates into ice cold 100% ethanol for 30 sec to fix the denatured DNA prior to centrifugation. Alternatively chemical blocking procedure can be used: For the chemical blocking process, use 1 g succinic anhydride (Fluka) dissolved in 200 ml anhydrous 1,2-dichloroethane (DCE; Fluka). To this solution add 2.5 ml of N-methylimidazol (Fluka) and immediately react with the microarray. Incubate 1 hour with slight agitation. Then wash the microarrays in fresh DCE and incubate in distilled water for 1 min for DNA denaturation. After a brief rinse in 95% ice cold ethanol to fix and dry.
5. Hybridized labeled samples prepared from DNA, mRNA and protein sources. Depending on the application, DNA microarrays prepared on SuperAmine and SuperAmine 2 surfaces can be hybridized with fluorescent and non-fluorescent DNA, messenger RNA (mRNA) and protein probe mixtures. Direct and indirect fluorescent and non-fluorescent labeling techniques have been described extensively in the literature and a host of commercial labeling kits are available from many different vendors. ArrayIt® offers an extensive line of purification, amplification and labeling kits for DNA, mRNA and protein labeling applications. For mRNA purification, amplification and labeling, ArrayIt® offers the Micro-Total RNA Extraction Kit, MiniAmp mRNA Amplification Kit, Indirect Amino Allyl Fluorescent Labeling Kit, Universal Reference mRNA, and Green540 2 and Red640 2 Reactive Fluorescent Dyes. For protein labeling applications, ArrayIt® Green540 and Red640 Reactive Fluorescent Dyes work extremely well for labeling. Labeled samples should be purified prior to microarray reactions to remove unincorporated dyes and other contaminants that can lead to uneven signals and elevated background. For hybridization reactions, ArrayIt® offers an excellent line of Hybridization Buffers for cDNA and oligonucleotide applications. Pre-heating the microarray substrate and the probe mixture to 42-65°C just prior to hybridization can greatly reduce background. For nucleic acid reactions, buffers typically contain 5X SSC or 5X SSPE and 0.1% detergent and incubations are typically preformed at 37°-65°C depending on the length of the targets and probes. ArrayIt® Hybridization Cassettes, available in various configurations and colors, provide excellent reaction environments for both DNA and protein microarray applications. The ArrayIt® Hybridization Station (Cat. HSAW), based on acoustic wave technology, enables automated hybridization. Following the microarray reaction, unbound probe material should be removed by successive washes in dilute buffers at room temperature (20-25°C). DNA and protein microarray applications typically use SSC or SSPE (DNA) and PBS buffers (protein), respectively.
6. Scan the microarrays to acquire data files. Once the microarrays are reacted with labeled samples and washed, the chips need to be scanned to produce images and data files. The ArrayIt® SpotLight scanners offer multi-color scanning at an affordable price. Additional systems are available from high quality vendors including Bio-Rad, Perkin Elmer, Axon Instruments (Molecular Devices), Applied Precision, Tecan, Alpha Innotech, and others. Scanners and imagers should be adjusted to select the appropriate detection wavelengths, resolution, gain, scan area, and other technical parameters to produce high quality images with maximum data content. Users should avoid excessive signal saturation, as this will “compress” the high intensity signals and cause a loss of dynamic range. In terms of scanning resolution, the pixel size should be approximately 1/10th the spot diameter such that 10 µm resolution would be ideal for microarrays containing 100 µm spots. Data files should be saved as 16-bit TIFF images for data quantification.
7. Perform data analysis. Scanned images contain microarray data as two-dimensional files with intensity values ranging from 0-65,536 (16-bit data). Mean and median spot signal intensities, normalized values, gene clusters, principal components analysis, and other statistical and computational approaches can be generated using commercial products offered by a number of vendors including BioDiscovery, PerkinElmer, SpotFire, Axon Instruments (Molecular Devices), VizXlabs, and others.

Figure 4. Shown is a photograph of a laser-etched fiducial mark on a SuperClean 2 Microarray Substrate. ArrayIt® has laser-etching capabilities suitable for making custom fiducials of any shape and dimension on SuperAmine and SuperAmine 2. Please inquire about our custom fiducials by electronic mail (arrayit@arrayit.com). The space bar corresponds to 100 µm (0.0039”).

Figure 5. Shown is a photograph of ArrayIt® H25K Human Genome, Cancer Splice Variant, Hematome and CheckIt Chips microarrays. Spots are visible due to reflection off the salt crystals formed in the printed elements. The SuperAmine and SuperAmine 2 surfaces provide the ultimate starting point for content microarrays, enabling high coupling efficiency, excellent spot morphology, high signals and low background.
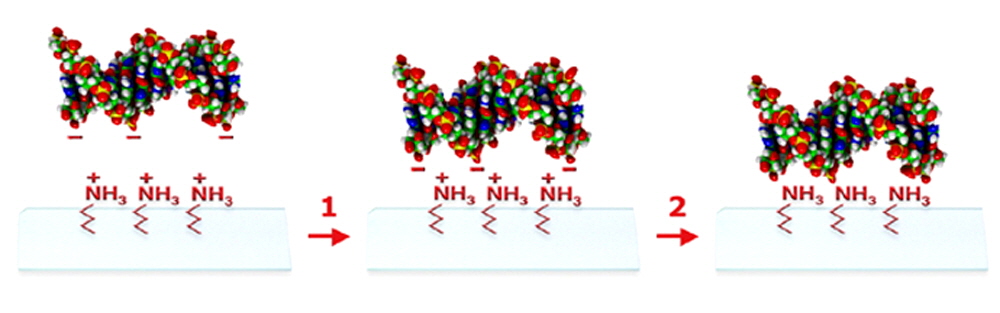
Figure 6. Shown is a schematic illustration of the SuperAmine 2 coupling mechanism. A negatively charged DNA molecule (space-filling model) is applied to the SuperAmine 2 surface using contact or non-contact printing (left panel). The printed sample dehydrates causing a loss of water and an large increase in the DNA concentration, resulting in attachment of the DNA to the SuperAmine 2 surface via the formation of electrostatic bonds (step 1) between the positively charged (+) amine groups and the negatively charged (-) DNA. The attachment of the DNA can be enhanced by the formation of covalent bonds (step 2) by baking at elevated temperature or treatment with ultraviolet light. Other types of negatively charged molecules can also be coupled to SuperAmine 2.
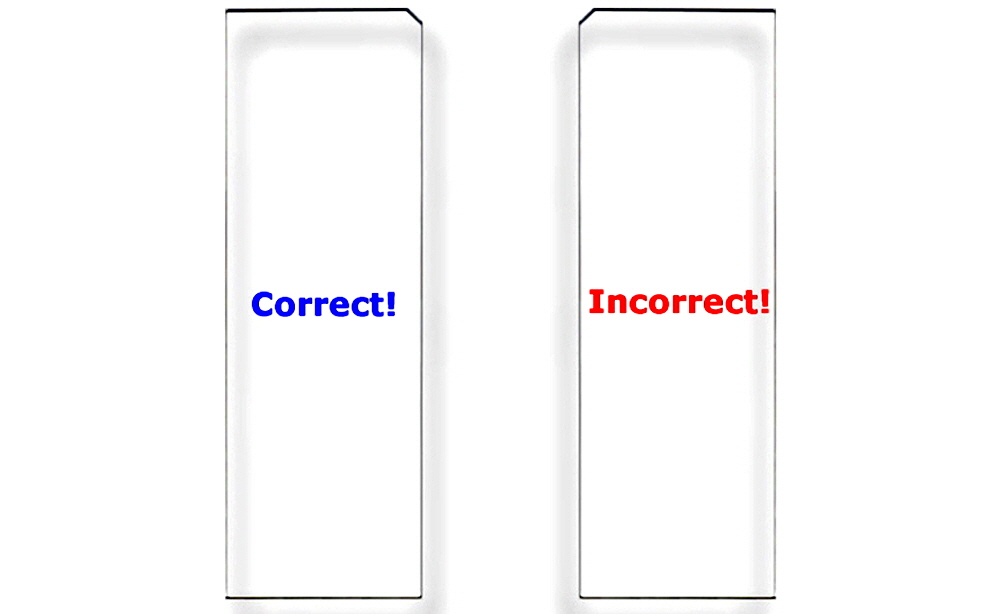
Figure 7. Correct Substrate Orientation. Shown is a graphic of two ArrayIt® Microarray Substrates, showing the correct and incorrect orientation for use. In the correct orientation (blue graphic), the chamfer will be located in the upper right corner and samples should be printed on the side facing upward, which is the same side that contains the word “Correct!”. In the incorrect orientation (red graphic), the chamfer will be located in the upper left corner, placing the backside facing upward, which is the side that contains the word “Incorrect!”. Only one side of ArrayIt® Microarray Substrates is suitable for printing. Please print on the correct side only.
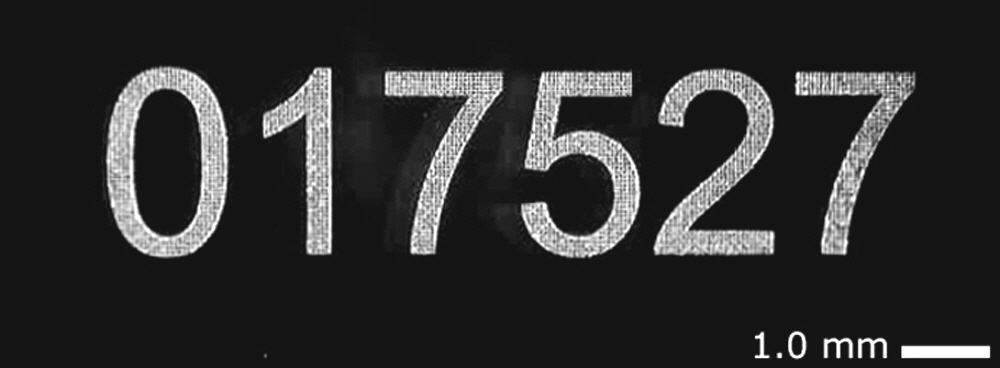
Figure 8. Laser-etched alpha numeric barcode. Photograph of a high-resolution 6-digit barcode etched at 1 µm precision into an atomically smooth 25 x 76 mm Arrayit microarray substrate slide. The space bar = 1.0 millimeter. Unlike adhesive labels, laser etching is resistant to the even harshest chemical and thermal treatments for the ultimate in sample tracking and identification purposes for diagnostics and other commercial applications. Barcodes, company logos, and custom graphics are available. Please contact arrayit@arrayit.com to place your order.
Technical Assistance
Please contact us if you have any questions, comments, suggestions, or if you need technical assistance. By electronic mail, use arrayit@arrayit.com and type "ArrayIt technical assistance" into the subject line. By email, arrayit@arrayit.com between the hours of 8AM and 7PM PST Monday through Friday. We want to hear about your successes and are always happy to publish contributed sample data on our website.
Troubleshooting Tips
1. Weak hybridization signals
- Impure samples. Need to purify cDNAs or oligos.
- Sample DNA concentration too low.
- Poor coupling. Make sure spots are dry and heat- or UV-treated.
- Make sure cDNAs are denatured prior to hybridization.
- Poor probe labeling. Check labeled products by gel or other means.
2. High background fluorescence
- Probe mixture drying out during hybridization. Keep hydrated.
- Probe volume too low. Use ≥2.0 µl per cm2 of cover slip.
- Poor processing or blocking. Follow Step 4 above.
- Probe mixture contaminated with unincorporated fluors. Need to purify.
- Hybridization time too long. Use a maximum time of 1-4 hrs.
- Poor washing procedure. Increase time or stringency.
Scientific Publications
Click here and here for recent scientific publications using ArrayIt® brand SuperAmine Microarray Substrates from Arrayit International, Inc.
Recommended Equipment and Reagents
NanoPrint™ 2 Microarrayers
SpotBot® 4 Personal Microarrayers
InnoScan® Microarray Scanners
Microarray Hybridization Cassettes
High Throughput Wash Station
Microarray High-Speed Centrifuge
Protein Printing Buffer
BlockIt™ Blocking Buffer
Microarray Air Jet
Microarray Cleanroom Wipes
PCR Purification Kits
Micro-Total RNA Extraction Kit
MiniAmp mRNA Amplification Kit
Indirect Amino Allyl Fluorescent Labeling Kit
Universal Reference mRNA
Green540 and Red640 Reactive Fluorescent Dyes
Hybridization Buffers

