SuperAldehyde Slides
Data Sheet
![]() Shop this product in our online store
Shop this product in our online store
Products - Microarray Substrates & Slides - SuperAldehyde and SuperAldehyde 2 DNA and Protein Microarray Glass Substrate Slides

ArrayIt® is pleased to announce SuperAldehyde 2, the newest version of our original SuperAldehyde Microarray Substrates product line. ArrayIt® substrates are the only microarray substrates in the world that offer a polished atomically smooth glass surface (±20 angstroms), for the ultimate in silicon dioxide homogeneity and data precision. SuperAldehyde 2 Substrates are manufactured to “open platform” dimensions (25 x 76 mm), cleaned at the atomic level in state-of-the-art class 1 cleanrooms, and treated with our ultra-pure aldehyde surface chemistry for superior covalent coupling efficiency and low background. Heat-sealing in anti-static packaging improves shelf life and eliminates electrostatic accumulation. SuperAldehyde 2 Substrates have a proprietary corner chamfer for unambiguous side and end orientation, which greatly improves usability during manufacturing. SuperAldehyde 2 products outperform standard optical quality glass from other vendors and are the substrates of choice for high-density DNA and protein microarray manufacturing applications using contact and non-contact printing methods. SuperAldehyde 2 products are compatible with all major brands of microarrayers and scanners.
Table of Contents
- Introduction
- Quality Control
- Product Description
- Technical Notes
- Glass Specifications
- Technical Assistance
- Short Protocols
- Complete Protocols
- Protein SuperAldehyde Short Protocol
- Protein SuperAldehyde Complete Protocol
- Recommended Equipment Requirements
- Troubleshooting Tips
- Ordering Information
- Storage Conditions
- Warranty
Introduction
Congratulations on taking a big step towards improving the affordability, quality and speed of your genomics, biomedical, pharmaceutical and agricultural research. This booklet contains all the information required to take full advantage of Arrayit's ArrayIt® Brand Super Microarray Substrates.
Quality Control
Arrayit takes every measure to assure the quality of our ArrayIt® Brand Super Microarray Substrates. The finest microarray biochip cleanroom research was used to develop these products. Rigorous quality control monitoring on a substrate-by-substrate basis guarantees that each these products conforms to the highest industry standards.
Product Description
Arrayit's ArrayIt® Brand Super Microarray Substrates provide the highest quality glass microarray printing substrates at an affordable price. All of our substrates are manufactured in state-of-the art class 1 cleanrooms, with 0.1 µm filtered air, and temperature and humidity control. This particle-free, sterile setting provides a quality of custom microarray substrates that cannot be obtained by any other vendor.
Users will appreciate the following:
- Polished to atomic surface smoothness <±20 angstroms over 1.0 µm2
- Superior substrate flatness of <0.036 mm over 25 x 76 mm
- Only polished glass substrate/slide in the microarray industry
- Homogenous SiO2 groups provide superior silane reactivity
- Reactive aldehyde density of 1 x 10^13 groups per mm^2.
- Vastly superior to unpolished optical quality glass from other vendors
- Ultimate surface for ultra-high density DNA and protein microarray manufacturing including micro-mirror, photolithography and contact printing
- Used by high-volume microarray manufacturing companies
- Used by overseas companies for diagnostics
- Topological smoothness ensures uniform hybridization layer and scanning
- Manufactured in a state-of-the-art class 1 cleanrooms
- Ultra-low intrinsic fluorescence and background noise
- Can be used as a starting point for gel and membrane coatings (e.g. polyacrylamide and nitrocellulose)
- Used by major academic core facilities
- Open platform dimensions compatible with all major brands of microarrayers and scanners
- Precise physical dimensions (25 ± 0.2 mm x 76 ± 0.3 mm x 0.940 mm ± 0.025 mm)
- Proprietary corner chamfer (1.4 mm) provides unambiguous side and end orientation to simplify printing and processing
- Finished edges enhance user safety
- Excellent refractive index, transmission and hardness specifications
- Sophisticated anti-static packaging improves usability
- Offered with or without bar-codes
- Custom laser and chrome fiducials available upon request
- Product arrives “ready to print” with no additional processing required
- High-volume 100,000 piece per month manufacturing capabilities
Technical notes
The “substrate noise” is the sum of all non-sample and non-instrument contributions to the background reading including intrinsic fluorescence of the substrate and reflection off the substrate surface. Because substrate noise is measured using a scanning or imaging device, as a practical matter substrate noise typically includes intrinsic fluorescence and reflection, as well as all of the sources of instrument noise (e.g. dark current, shot noise, electronic noise and optical noise). Most modern scanners and imagers have very low instrument noise, which means that intrinsic fluorescence and reflection dominate the substrate noise reading. Substrate noise is obtained by measuring the fluorescent reading of the substrate “right out of the box” and prior to reacting the substrate with a fluorescent sample. Please do not confuse substrate noise (i.e. background before the substrate is reacted with a sample) and microarray noise (i.e. background after the substrate has been reacted with a sample), as these two readings are very different. For nearly all applications and assays, microarray noise greatly exceeds substrate noise and therefore concerns about substrate noise are entirely academic because the latter does not contribute to the total background reading observed when the product is used in real experiments (see below).
The table below summarizes the substrate noise (actually intrinsic fluorescence + reflection + instrument noise) observed with our substrates. Substrate noise readings were taken at very high sensitivity (90% laser and 90% photomultiplier tube or PMT), settings that are well beyond those used for biological experiments. Typical instrument settings for biological experiments with the ScanArray Express are 20-40% laser power and 80% PMT. The ScanArray Express has 20- to 30-fold greater sensitivity than the ScanArray 3000, and correspondingly the substrate noise readings are up to 30-fold higher on the Express compared to the 3000. Unmodified glass substrates (i.e. SuperClean) produce the lowest substrate noise readings, followed SuperEpoxy, SuperAmine, and SuperAldehyde. Organic treatments (e.g. epoxy, amine and aldehyde) increase substrate noise compared to naked glass because organic molecules formed during derivation fluoresce at an extremely low but detectable level. For nearly all applications and assays, the non-sample contributions to background noise (intrinsic fluorescence, reflection, and instrument noise) are much less than the background noise contributed when the substrate or microarray is reacted with a fluorescent sample. In these cases, substrate noise (though it exists) is irrelevant to the use of our products, because it does not contribute in any measurable way to the background reading of the reacted chip. In rare cases involving extremely low background samples or gene expression measurements of rare transcripts, substrate noise may approach sample noise in magnitude.
For best results, please test our products in the context of REAL EXPERIMENTS rather than simply taking note of the fact that our Substrates manifest substrate noise that is greater than plain glass. ALL SUBSTRATES that contain an organic treatment or coating will produce some intrinsic fluorescence and reflection. Please also test our product “right out of the box” rather than waiting hours or days to measure the substrate noise. Fluorescent contaminants present in non-cleanroom air including cleaning agents, solvents used in marking pens, and hydrocarbon emissions from vacuum pumps, arrayers, centrifuges and other instruments can elevate the substrate noise reading considerably. Please also note that airborne particles including dust and other particulates greatly elevate the background reading because these particles are highly reflective in the presence of laser and white excitation light. Understanding the technical details of our products is important and we recommend that you commit these concepts to working memory as you proceed with your experiments.
Table 1. Substrate noise
Product |
Average substrate noise (ScanArray 3000) |
Maximum allowable substrate noise (ScanArray 3000) |
Average substrate noise (ScanArray Express) |
Maximum allowable substrate noise (ScanArray Express) |
SuperAldehyde |
278 |
500 |
729 |
2,000 |
MirrorAldehyde |
217 |
375 |
568 |
1,500 |
Glass Description and Specifications:
The glass used for these substrates has been especially manufactured exclusively for this application. These are not treated microscope slides! Glass substrates of this optical quality and smoothness are sold for up to $300.00 dollars a piece by vendors in the scientific glass industry.
1. Standard substrate format (25 mm x 76 mm x 0.960 mm).
2. Chamfer at upper right corner allows unambiguous substrate orientation.
3. Polished to atomic smoothness of <50 Å.
4. Low refractive index (400-700 nm).
5. High light transmission efficiency (380-700 nm).
6. Proprietary glass formulation much harder than normal glass.
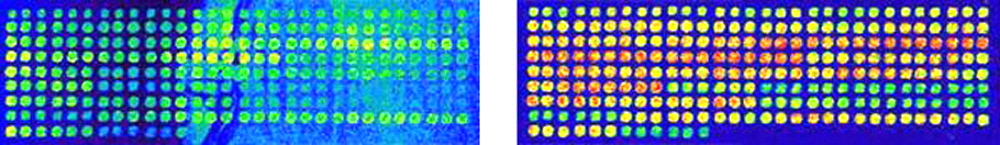
Figure 1. Improved hybridization results with probe pre-heating and BSA addition. Shown are two microarrays of PCR products printed on SuperAldehyde Substrates (Arrayit) with Stealth Micro Spotting Pins (SMP3B) and a Cartesian 5500 robot. Printed microarrays were washed to remove unbound material and the double-stranded DNAs were denatured by boiling for 3 min in distilled water. The microarrays were hybridized in hybridization cassettes (Arrayit) for 5 hrs at 42°C under 18 mm x 18 mm optically flat cover slips with 5.0 µl probe solution containing 5X SSC + 0.2% SDS + 0.2 mg/ml bovine serum albumin (BSA) + 2 µM 15-mer oligonucleotide. The 15-mer oligonucleotide contained a Cy3 label on the 5' end. Hybridized microarrays were washed twice for 5 min each in 2X SSC + 0.2% SDS at 25°C, once for 1 min in 2X SSC at 25°C and spun dry for 1 min at 500 x g. Microarrays were scanned at 100% photomultiplier tube (PMT) and 100% laser settings with a ScanArray 3000 (Packard Instruments). (Left) Probe solution at 25°C applied to microarray and (Right) Probe solution pre-heated to 42°C and applied IMMEDIATELY to microarray. Improved results are easily observed in the right image compared to left image, including stronger signals, reduced background and increased uniformity. Probe pre-heating and BSA addition can both improve microarray data.

Figure 2. Atomic Force Microscopy (AFM) analysis. Shown are AFM scans for SuperClean (top), SuperAmine (middle) and SuperAldehyde (bottom) Microarray substrates. Data are coded to a rainbow intensity scale, such that red data represents 5.0 nm or 50 angstroms. The substrates have an average smoothness of 2.0 nm or 20 angstroms, which corresponds to the about 10 silicon dioxide bonds.
Technical Assistance
Please contact us if you have any comments, suggestions, or if you need technical assistance. By electronic mail: arrayit@arrayit.com (under the subject heading please type "ArrayIt® technical assistance"). By email: arrayit@arrayit.com, Monday–Friday PST 9:00am - 4:30pm. Please remember that we want to hear about your successes!
Short Protocol (SuperAldehyde)
1. (Optional) Use an oil-free air stream to remove particles from plastic packaging exterior.
2. (Optional) Transport de-contaminated product into cleanroom.
3. Remove plastic packaging and discard.
4. Open substrate box to reveal 25 SuperAldehyde Microarray Substrates.
5. Print microarrays using either contact printing or ink-jet technology.
6. Process the printed microarrays.
7. React printed microarray with a labeled sample.
8. Image microarray with a fluorescence detection device.
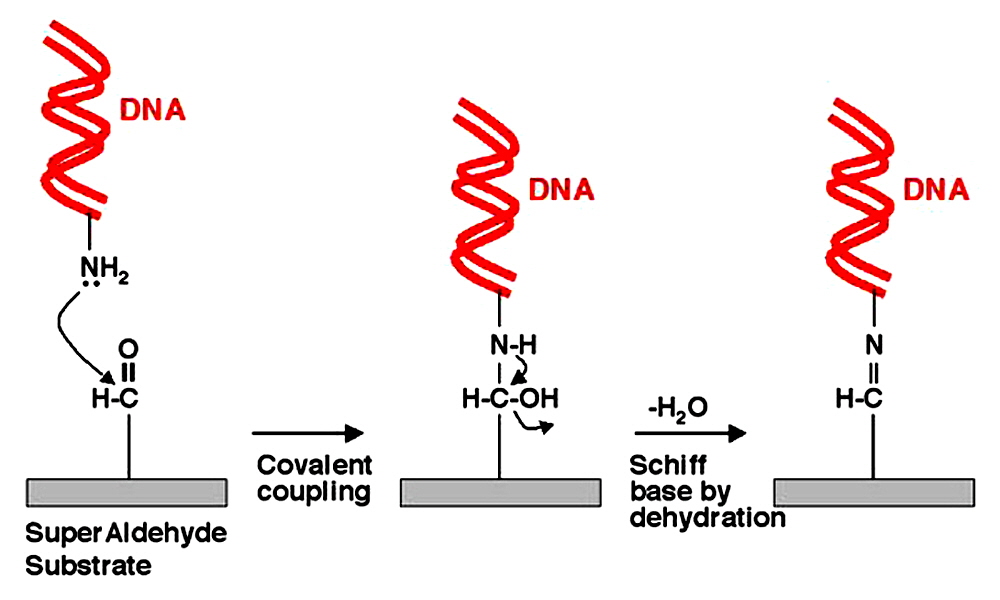
Figure 3. SuperAldehyde Coupling Chemistry. SuperAldehyde Substrates contain primary aldehyde groups attached covalently to the glass surface (rectangles). Primary amino linkers (NH2) on the DNA (red ribbons) attack the aldehyde groups (left panel) forming covalent bonds (center panel). Attachment is stabilized by a dehydration reaction (drying in low humidity) which leads to Schiff base formation (right panel). Specific and covalent end attachment provide highly stable and accessible attachment of DNA for gene expression and genotyping applications. Proteins can also be attached to SuperAldehyde, though the SuperEpoxy surface is generally preferred for protein microarrays.
Complete Protocol (SuperAldehyde)
1. (Optional) Use an oil-free air stream to remove particles from the plastic packaging exterior. This step is optional and only required for users who fabricate microarrays in a cleanroom environment. Shipping material can contaminate the exterior packaging material with particulates that can be transferred into the cleanroom. These particulates can be removed from the sealed plastic shipping envelope using a stream of pressurized air from an oil-free source. A variety of oil-free air compressors (e.g. Sears) work well for this purpose.
2. (Optional) Transport the de-contaminated product into the cleanroom. This step is optional and only required for users who fabricate microarrays in a cleanroom environment. Carry the de-contaminated SuperAldehyde Microarray Substrates in the sealed plastic shipping envelope into the cleanroom changing area. Change into cleanroom attire and enter the cleanroom facility.
3. Remove the plastic packaging envelope and discard it into an appropriate waste receptacle. The plastic shipping envelope should be opened and discarded at a location distant from the printing area, to prevent transfer of residual contaminants onto the printing surface.
4. Open the substrate box to reveal 25 SuperAldehyde Microarray Substrates. Use the notch at the base of the Substrate box to gently lift the cover upward. Do not attempt to open the Substrate box upside down as this will cause the SuperAldehyde Substrates to empty into the lid of the box and may damage or break the glass.
5. Print the microarrays using either contact printing or ink-jet technology. Load the SuperAldehyde Microarray Substrates onto the printing surface of a microarraying device, with the corner chamfer located at the upper right corner as shown in Figure 2. Because the SuperAldehyde coating is applied only to the top side of the Substrate, DO NOT attempt to print on the non-coated, back side of the Substrate. The printing convention places spot number one in the upper left corner opposite the corner chamfer (see Figure 2). Print the microarrays until all of the samples have been deposited. The maximum recommended printing area is 20 mm x 72 mm.
6. Chemically couple the biomolecules to the SuperAldehyde surface, process the printed microarrays to remove unbound material, and block unreacted aldehyde groups to reduce non-specific binding of labeled reactants. Optimal protocols for proteins, small molecules, extracts, cells and other molecules have been developed. One protocol that works well for printing and attaching single- and double-stranded nucleic acids to the SuperAldehyde surface uses DNA modified with primary amines that form Schiff base covalent bonds with reactive aldehydes on the Substrate surface. (1) attach a C6 amino modification (Glen Research) to the 5' end of each oligonucleotide; amino modifications can be added to double stranded DNAs by PCR with primers that contain amino modifications, (2) re-suspend the amino-linked DNAs in 1X Micro Spotting Solution Plus, (3) print the amino-linked DNAs onto SuperAldehyde Microarray slide Substrates with Stealth Micro Spotting Pins, (4) allow the Substrates to dry for 12 hrs at room temperature (~25°C) at <30% relative humidity, (5) rinse the printed substrates twice in 0.1% SDS and once in dH2O for 2 min each wash at room temperature (22-25°C) to remove unbound DNA, (6) rinse the Substrates once in dH2O for 2 min at room temperature with vigorous agitation, (7) transfer the Substrates into boiling dH2O at 100°C for 3 min to denature the DNA (8) plunge the substrates into ice cold 100% ethanol for 30 sec to fix the denatured DNA, and (9) dry the processed Substrates by centrifugation for 10 sec in a Microarray High Speed Centrifuge or for 1 min at 500 x g in a traditional centrifuge (e.g Savant). A 1 hour pre-hybridization treatment with BlockIt buffer prior to hybridization can be used to reduce background in all fluorescent DNA and protein based assays. Additionally, for DNA microarrays only, treatment with sodium borohydride (NaBH4) can be used to reduce unreacted aldehyde groups to alcohols. This can lower fluorescent background in some assays. For sodium borohydride reduction, follow Steps 1-6 exactly as written above, and then proceed as follows: (7) reduce for 5 min with gentle mixing in 500 ml sodium borohydride reducing solution [dissolve 1.5 g NaBH4 in 350 ml phosphate buffered saline (PBS) and add 150 ml 100% ethanol after the NaBH4 dissolves to improve reducing efficiency], (8) rinse 2 times for 1 min each in 500 ml dH2O, (9) rinse 2 times for 1 min each in 500 ml 0.1% SDS, (10) transfer the Substrates into boiling dH2O at 100°C for 3 min to denature the DNA (11) plunge the substrates into ice cold 100% ethanol for 30 sec to fix the denatured DNA, and (12) dry the processed Substrates by centrifugation for 10 sec in a Microarray High Speed Centrifuge or for 1 min at 500 x g in a traditional centrifuge (e.g Savant). High Throughput Wash Stations work well for all wash steps except those involving elevated temperatures (e.g. boiling).
7. React the processed microarray with a labeled probe sample. Probes can be complex mixtures of labeled or fluorescent nucleic acids, proteins, antibodies, extracts and other substances. Microarray reactions are best performed under glass cover slips at a volume of 2.0 µl per cm2 of cover slip. Pre-heating the probe to 42-65°C just prior to hybridization can greatly reduce background!! For nucleic acid reactions, buffers typically contain 5X SSC or 6X SSPE and 0.1% SDS and are preformed at 37°-65°C depending on the nature of the targets and probes. The addition of 0.2 mg/ml BSA (Worthington) to the hybridization reaction reduces background. UniHyb Hybridization Solution (Arrayit) also works well at a wide range of temperatures and reduces background hybridization is many cases. Hybridization Cassettes(Arrayit) provide a convenient reaction environment, automation can also be used. Following the microarray reaction unbound probe material is removed by several successive washes in dilute buffers at room temperature (~25°C).
8. Image the microarray with a fluorescence detection device. Suitable detection systems accommodate the 25 mm x 76 mm (1" x 3") substrate and have a pixel resolution corresponding to ~1/10 the diameter of each microarray feature. Popular detection systems are available from Arrayit.
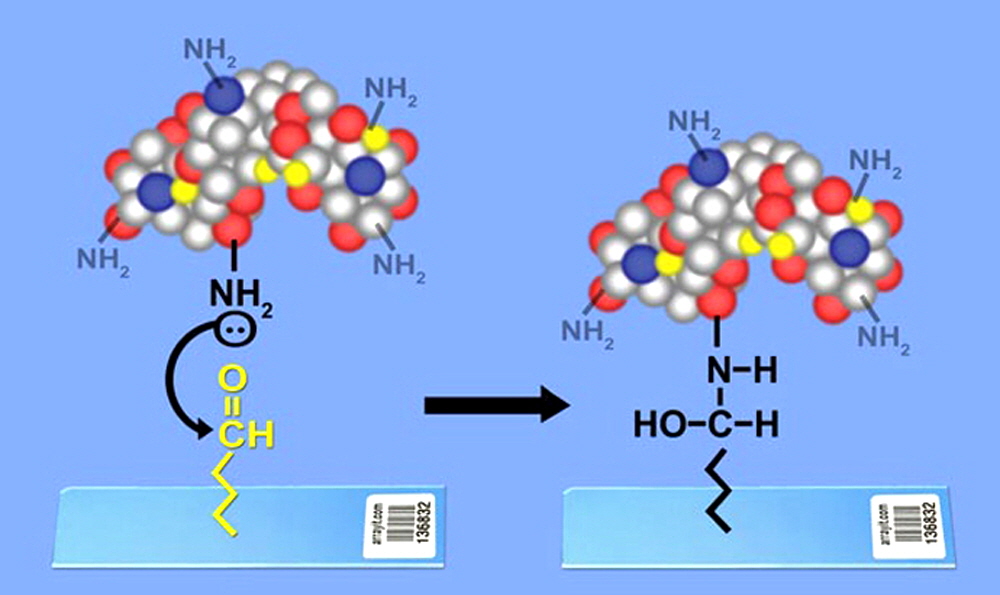
Figure 4. Protein coupling to the SuperAldehyde surface.SuperAldehyde Substrates contain primary aldehyde groups attached covalently to the glass surface. Primary amino linkers (NH2) on proteins attack the aldehyde groups (left) forming covalent bonds (right panel). Attachment is stabilized by a dehydration reaction (drying in low humidity) that leads to Schiff base formation. Covalent attachment provides highly stable and accessible attachment of protein for various protein assays including protein-protein interactions studies, protein profiling, and sandwich assays. Proteins can also be attached to SuperEpoxy.
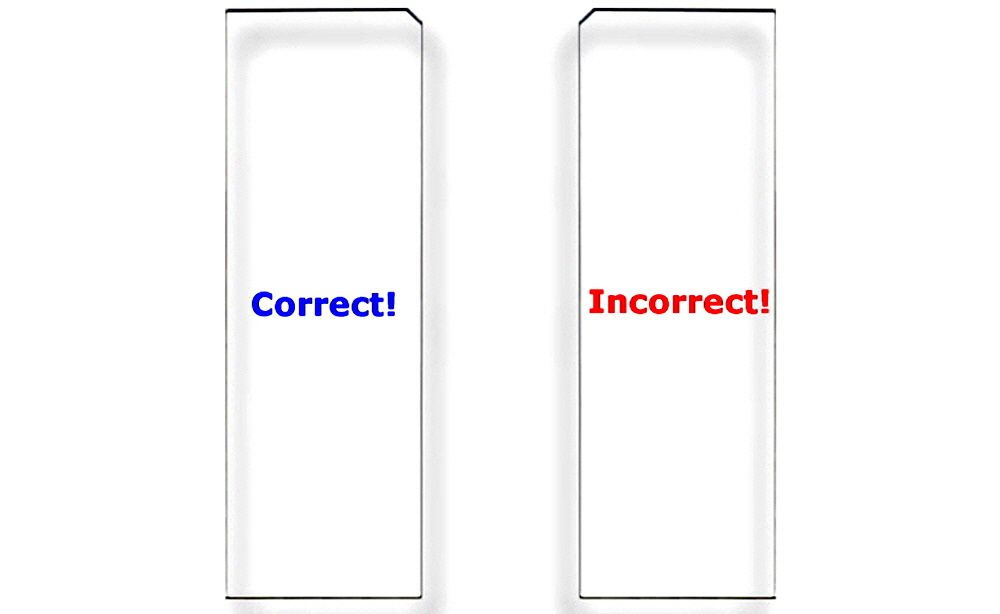
|Figure 5. Correct Substrate Orientation. Shown is a graphic of two ArrayIt® Microarray Substrates, showing the correct and incorrect orientation for use. In the correct orientation (blue graphic), the chamfer will be located in the upper right corner and samples should be printed on the side facing upward, which is the same side that contains the word “Correct!”. In the incorrect orientation (red graphic), the chamfer will be located in the upper left corner, placing the backside facing upward, which is the side that contains the word “Incorrect!”. Only one side of ArrayIt® Microarray Substrates is suitable for printing. Please print on the correct side only.
Figure 6. SuperAldehyde 2 surface chemistry can be purchased in the “SuperMask” hydrophobic mask format. Custom patterns are available.
SuperAldehyde Protein Microarray Short Protocol (Steps 1-7):
1. Suspend protein samples in Protein Printing Buffer at 0.1-0.5 µg/µl.
2. Print protein samples onto SuperAldehyde Substrates.
3. Block using BlockIt buffer and process the microarrays.
4. React the processed microarrays with fluorescent samples.
5. Wash the microarrays to remove unreacted fluorescent material.
6. Scan the microarray to produce a fluorescent image.
7. Quantitate and model the fluorescent data.
SuperAldehyde Protein Microarray Complete Protocol (Steps 1-7):
1. Suspend protein samples in Protein Printing Buffer. Obtain 0.2-1.0 µg/µl protein samples in 1X phosphate buffered saline (PBS) and add an equal volume of 2X Protein Printing Buffer. Mix the samples by pipetting up and down 10 times. Protein samples should be free of aggregates and particulates that can clog printing devices and impair attachment to the microarray substrate. Aggregates and particulates can be removed by centrifugation or filtration. A 50kD protein at 1 µg/µl concentration has a concentration of 20 µM. At 30% coupling efficiency, a 20 µM protein will produce a target density of 1011 proteins per mm2 of substrate. Certain proteins or protein extracts are more stable at 4°C. Keeping the protein samples cool may improve protein stability. Stability can also be improved in some cases by the addition of protease and phosphatase inhibitors, or by the use of a SpotBot Personal Microarrayer or NanoPrint Microarrayer equipped with a cooled platen and/or plate cooling. Make sure protein samples are mixed thoroughly before printing.
2. Print protein samples onto SuperAldehyde Substrates. The surface couples proteins extremely efficiently owing to the high reactivity of the aldehyde groups to primary amines (see Fig. 4). To complete coupling of protein to the surface wait several hours to overnight after printing at low humidity prior to using the microarrays. Proteins retain protein-protein binding activity after the spots have dried. Printed microarrays should be stored unprocessed to protect coupled molecules. Protein Printing Buffer contains components that stabilize printed proteins. Processing should be performed just prior to use for best performance.
3. Block and process the printed protein microarrays. Once the printing process is complete, wash the printed microarrays to remove unbound protein molecules and buffer components from the surface. Protein binding to is extremely stable and the microarrays can be washed, blocked and reacted without sufficient loss of coupled protein. For best results block the surface for 1 hour using BlockIt blocking buffer. If BlockIt is not immediately available, a 1 hour incubation at room temperature in a buffer containing 1X PBS + 1% bovine serum albumin (BSA) can be used with satisfactory results. A High Throughput Wash Station or an equivalent device can be used for washes. Blocking can be performed with very gentle buffer agitation and the stir plate set on a low speed or under a coverslip. The blocking step will couple reactants to the aldehydes on the surface and prevent background fluorescence. After blocking, wash the microarrays to remove the excess blocking buffer. Washing three times for 2 min each at room temperature with 1X PBS in a High Throughput Wash Station works well.
4. React the processed microarrays with fluorescent samples. Processed microarrays containing coupled target proteins can be reacted with fluorescent samples to study protein-protein interactions or for gene expression monitoring. Binding reactions can be performed in 1X BlockIt buffer for best results. Alternatively 1X PBS + 0.5% BSA + fluorescent proteins diluted 1:1,000 or labeled cellular extracts. Fluorescent samples can be incubated as a droplet on the printed microarray, underneath a cover slip, or in a microfluidics chamber. A 60-minute incubation at room temperature is usually sufficient to obtain strong binding and intense fluorescent signals. A Hybridization Cassette can be used to prevent sample evaporation during prolonged (1-12 hour) binding reactions.
5. Wash the microarrays to remove un-reacted fluorescent material. Once the binding reaction between the bound target proteins and the fluorescent protein probe molecules is complete, wash the microarray to remove the unbound fluorescent material. Washes can be performed three times for 5 min each at room temperature in 1X PBS. After the wash procedure, excess buffer should be removed from the surface by tapping the substrate on a lint-free cloth or by centrifugation with a Microarray High-Speed Centrifuge.
6. Scan the microarray to produce a fluorescent image. The fluorescent protein microarray can be scanned or imaging using any of a number of high quality commercial detection instruments from Perkin Elmer, BioRad, Axon , API and many others. Instrument settings can be adjusted to optimize the imagine process.
7. Quantitate and model the fluorescent data. Protein microarray data from the fluorescent image can be quantified, mined and modeled using many different commercial software packages including BioDiscovery (Marina del Ray, CA).
Troubleshooting Tips
1. Weak signal
· Poor coupling of printed material to Substrate
· Poor labeling of probe sample
2. High background fluorescence
· Drying of labeled sample during reaction or washes
· improper reaction or wash conditions
 SuperAldehyde substrates are offered with barcodes:
SuperAldehyde substrates are offered with barcodes:
ArrayIt® barcodes are 12 mm x 19 mm and are affixed at the bottom edge of the 25 mm x 76 mm substrate (smaller custom barcodes are available upon request).
Scientific Publications
Click here and here for recent scientific publications using ArrayIt® brand SuperAldehyde Microarray Substrates from Arrayit International, Inc.
Recommended Equipment and Reagents
NanoPrint™ 2 Microarrayers
SpotBot® 4 Personal Microarrayers
InnoScan® Microarray Scanners
SpotLight™ 2 Microarray Scanners
Microarray Hybridization Cassettes
High Throughput Wash Station
Microarray High-Speed Centrifuge
Protein Printing Buffer
BlockIt™ Blocking Buffer
Microarray Air Jet
Microarray Cleanroom Wipes
PCR Purification Kits
Micro-Total RNA Extraction Kit
MiniAmp mRNA Amplification Kit
Indirect Amino Allyl Fluorescent Labeling Kit
Universal Reference mRNA
Green540 and Red640 Reactive Fluorescent Dyes
Hybridization Buffers

