Universal RNAs
Data Sheet
![]() ArrayIt® Rat Universal Reference mRNA
ArrayIt® Rat Universal Reference mRNA
![]() ArrayIt® Mouse Universal Reference mRNA
ArrayIt® Mouse Universal Reference mRNA
![]() ArrayIt® Human Universal Reference mRNA Cy3
ArrayIt® Human Universal Reference mRNA Cy3
![]() ArrayIt® Human Universal Reference mRNA Cy5
ArrayIt® Human Universal Reference mRNA Cy5
Arrayit | Universal ribonucleic acid RNA human mouse rat microarray control expression profiling life sciences research
Reagents - Microarray Labeling - Reference mRNAs & Labeled cDNA Mixtures for DNA Microarray Applications in Gene Expression Profiling, SNP Analysis and Genomics
Universal Reference Human mRNA is a proprietary blend of amplified high-quality mRNA purified from a diverse set of human, mouse or rat tissues. This reference mRNA provides a common denominator for accurate and reproducible comparisons of gene expression data. Pooled reference mRNA can be used for multicolor hybridization experiments using DNA microarrays. The signal generated from this mRNA will ease the normalization process, in both intensity-based and reference-channel-based normalization. The use of pooled reference RNA has been shown to reduce inter and intra-laboratory variations of microarray data. This product is compatible with all commercially available microarray labeling systems as well as other research applications such as RT-PCR, and QRT-PCR that demand high quality reference pool of mRNA. New Universal Reference Cy3- and Cy5-labeled cDNA mixtures offer exceptional normalization standards for two-color hybridization experiments.
Reference mRNA can also be delivered as indirectly labeled cDNA (see ordering information).
Can be used stand alone or in conjunction with:
- Indirect Amino Allyl Fluorescent Labeling Kit
- Micro-Total RNA Extraction Kit
- MiniAmp mRNA Amplification Kit
Uses include:
- Normalization standard for two-color microarray systems
- Standardization method for single color microarray systems
- Real-time PCR amplification efficiency and standardization studies
ArrayIt Universal Reference mRNA:
- Provides more broad gene representation over competing products
- Is available for three different species: Human, Mouse, and Rat
- Guaranteed high purity mRNA, free of contamination
- Bulk production minimizes lot-to-lot variation
- Highly useful for any DNA based array, microarray or PCR platform
Packaging:
- 30 µg total mRNA in a 0.5 ml micro-centrifuge tube, concentration 1µg/µl
- 2 µg labeled cDNA, dried down and packed under nitrogen in a light proof tube
Storage:
- Store at –80° C stable for 6 months at this temperature.
- It is advisable to minimize handling and repeated freeze-thaw of this product; we recommend that at the time of first use the product be split into smaller, single use size aliquots, using RNAse free tips and storage vials.
Differences between using total RNA and mRNA in labeling for microarrays are as follows:
1. When labeling using total RNA in either direct or in-direct methods, oligo dT to prime the RT reaction is used. Therefore the labeling is limited to the capability of the reverse transcriptase to generate large fragments on a single run. On the other hand, when using mRNA, a mixture of oligo dT and random primers is used, allowing a much better cDNA:dye ratio and much better labeling. For this reason, many microarray users prefer to purify mRNA prior to reverse transcription and labeling.
2. The use of reference mRNA becomes absolutely necessary and saves a lot of time and money, when a user is working from small quantities of RNA and is performing mRNA amplification for microarray studies. The amplified mRNA from the samples can be used directly with the reference mRNA, saving one round of amplification. Since, the sample total RNA (purified by the user) and the reference total RNA (purified by the vendor) will have different purity, the results will vary in the amplification reaction as well.
Number of labeling per pack of 30 µg
For direct labeling use 2-3 µg/reaction, for indirect labeling use 1-2 µg/reaction.
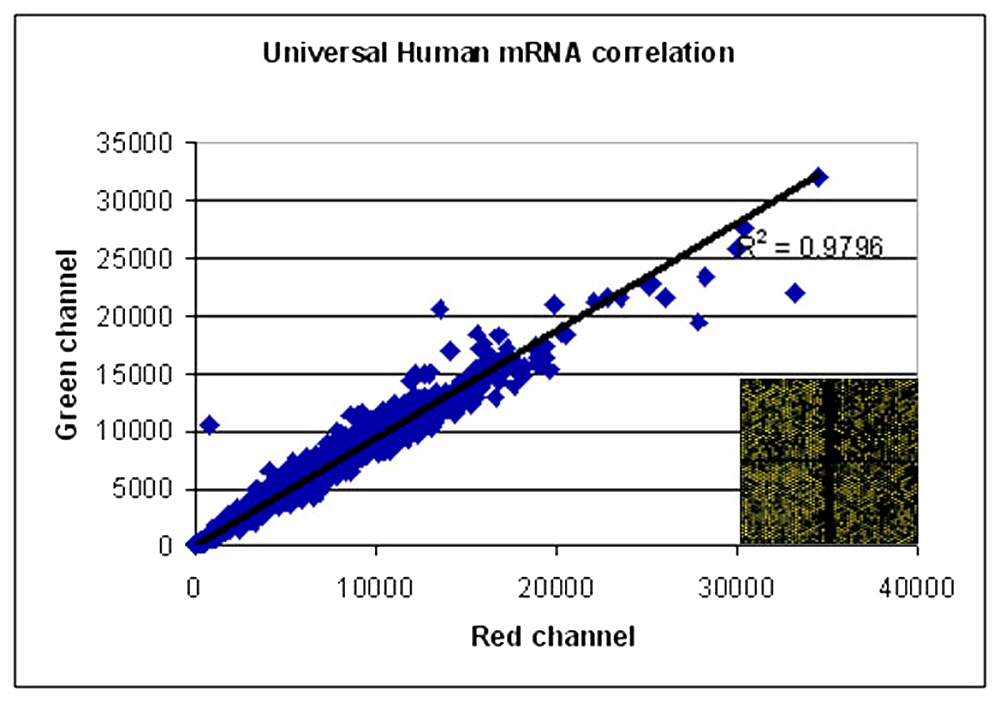
Figure 1. Quality testing of Human Universal mRNA
- Step 1. Two equal aliquots (2.5 µg) of Human universal mRNA was labeled with Cy3 and Cy5 labeled dCTP (Amersham) each using reverse transcriptase and anchored oligo dT primer and random nanomer.
- Step 2. After separating the bound and free dye the labeled cDNAs were mixed and hybridized to a cDNA microarray containing 9200 human ESTs
- Step 3. After washing the microarray was scanned using an Axon scanner
- Step 4. The scanned image was gridded with Axon Genepix software the results were subjected to the following analysis: Step 1A. The median value of (intensity – background) was taken and filtered using the criteria that Signal > Background + 2x SD of Background Step 2A. Those spots whose signal value was less than 100 in either of the channels were removed. Step 3A. After all these filtering 8120 spots or 88 % of the spots gave good results Step 4A. When a correlation graph was plotted for Red: Green channel values the correlation coefficient was 0.9796

