Hybridization Buffers
Data Sheet
![]() Shop this product in our online store
Shop this product in our online store
Arrayit | Microarray hybridization buffers solutions HybIt UniHyb formamide detergent salt kinetics DNA protein microarrays
Reagents - Microarray Buffers - Hybridization Buffers - HybIt® 2 Formamide-Based Hybridization Buffer for DNA Microarrays

Arrayit has developed HybIt® 2 Hybridization Solution containing an advanced formamide mixture that improves microarray hybridization reactions by increasing signal intensities and reducing background in reactions with cDNAs and long oligonucleotides. Use HybIt® to produce superior data with microarrays printed on atomically flat Super Microarray Glass Substrate Slides. HybIt® 2 Hybridization Solution Automation Formulation is highly recommended for use with TrayMix™ Hybridization Stations.
Table of Contents
Introduction
Quality Control
Product Description
Technical Assistance
Short Protocol
Complete Protocol
Requirements
Troubleshooting Tips
Ordering Information
Warranty
Introduction
Congratulations on taking a big step towards improving the economies of scale, quality and speed of your genomics research. This booklet contains a complete set of protocols outlining the steps and principles needed to use Arrayit HybIt® 2 Hybridization Solution.
Quality Control
Arrayit assures the performance of this product. The finest scientific research went into the development of this product. Rigorous quality control monitoring on a lot-by-lot basis guarantees that the ingredients conform to the highest industry standards.
Product Description
The Arrayit HybIt® 2 Hybridization Solution is an advanced hybridization solution containing a patent-pending mixture of salts, detergents and buffering components. Use of HybIt® 2 Hybridization Solution will increase the quality of microarray biochip hybridizations reactions involving base pairing interactions between complementary nucleic acid chains. Users will appreciate the following features:
- Increases signal by accelerating hybridization kinetics
- Increases sensitivity by reducing background fluorescence
- Reduces surface tension providing a uniform hybridization layer
- Buffering components stabilize extended reactions
- Compatible with many glass-based surface chemistries
- Arrives pre-mixed and sterile, no preparation required
- Costs <$1 dollar per hybridization reaction
Technical Assistance
Please contact us if you have any comments, suggestions, or if you need technical assistance. By electronic mail: arrayit@arrayit.com (under the subject heading, please type Arrayit technical assistance). By email: arrayit@arrayit.com, Monday–Friday PST 9:00am - 4:30pm. Please remember that we want to hear about your successes!
Short Protocol (Steps 1-7)
1. Print microarrays or obtain Arrayit Custom printed microarrays.
2. Process the microarrays for hybridization.
3. Purify probe using ArrayIt Fluorescent Probe Purification Kit.
3. Resuspend fluorescent probe in 1.0 part dH20 and 4.0 parts HybIt® 2 Solution.
3. Hybridize the probe to the microarray under the appropriate conditions.
5. Wash away the unbound fluorescent probe. ArrayIt wash buffers A, B & C are recommended.
6. Scan the microarray for fluorescent signal.
7. Score the hybridization results.
Complete Protocol (Steps 1-7)
1. Print microarrays with an Arrayit Micro Spotting device or a suitable microarray technology. In principle, HybIt® 2 should improve detection on many different microarray platforms.
2. Process the microarrays for hybridization. After the microarrays are spotted, the slides should be allowed to dry at room temperature overnight for arrays printed on SuperAldehyde. This can be accomplished by placing the slides in a slide box with the lid slightly ajar. Drying increases the binding of the amino-modified DNA to the aldehyde surface. After drying, the slides should be processed to remove unbound DNA. Many protocols have been used for slide processing. The hybridization section at http://arrayit.com for more information.. Microarrays printed on SuperAmine should be crosslinked by either baking at 80°C for 80 minutes or by using a UV Crosslinker (120 mJ).
3. Purify the fluorescent probe with the ArrayIt Fluorescent Probe Purification Kit and resuspend in HybIt® 2 Hybridization Solution, which is provided as a 1.25X solution. This is accomplished by first resuspending the probe in 1.0 part dH20, and then adding 4.0 parts of HybIt® 2 Hybridization Solution. Prior to using HybIt® 2 Hybridization Solution, pre-warm the solution for 30 sec at 42°C and mix by inverting the tube several times. A fluorescent probe desiccated to dryness would be re-suspended by adding 2.0 µl of dH20, followed by 8.0 µl of pre-warmed HybIt® 2 Hybridization Solution. Probes should be single-stranded DNA or RNA molecules made by either reverse transcription or in vitro transcription of cRNA.
4. Hybridize the probe to the microarray under the appropriate conditions (42°C). This is accomplished by using 1.25 µl of probe in 1X HybIt® 2 per cm2 glass cover slip. For best results, add the probe to one edge of the cover slip surface, and then gently lower the cover slip onto the microarray with fine forceps allowing the probe to sheet evenly across the surface between the cover slip and the slide. Transfer the substrate with cover slip to a pre-warmed Arrayit Hybridization Cassette containing 10.0-µl warmed dH20 in each well. Also use 10-20 ul of warmed dH20 under the substrate. Seal the cassette and hybridize for overnight at 42°C.
5. Wash away the unbound fluorescent probe. Remove the microarray from the Hybridization Cassette and immediately transfer the slide to an Arrayit Wash Station and wash according to directions.
6. Scan the microarray for fluorescent signals. Insert the substrate into the ScanArray 3000 (Packard Bio Science) or a compatible detection system and scan the area of the slide containing the microarray. The scan area, excitation source, laser power and PMT settings can all be adjusted with the ScanArray software. Laser and PMT settings should be chosen to give maximal unsaturated signal with minimal background fluorescence. Typically, laser and PMT settings of 70% and 60-80% respectively yield good results with the ScanArray 3000.
7. Score the hybridization results. Upload the scanned image tiff file into the ImaGene software (BioDiscovery) or a suitable quantitation package and examine each feature for fluorescence intensity.
Requirements
Micro Spotting Device
High Throughput Wash Station
Fluorescent Probe Purification Kit
Micro-Spotting Solution Plus
Hybridization Cassettes
Super Microarray Substrates
Troubleshooting Tips
High Background:
Impure cDNAs or oligonucleotides used for spotting.
Used conventional buffers instead of HybIt 2 Hybridization Solution.
Incorrect hybridization temperature.
Unincorporated fluors were not sufficiently purified away from probe, use Fluorescent Probe Purification Kit.
Reagents - Microarray Buffers - Hybridization Buffers - HybIt® Hybridization Solution for DNA Microarray Hybridization Reactions
Arrayit has developed HybIt® Hybridization Solution containing an advanced mixture that improves microarray hybridization reactions by increasing signal intensities and reducing background in reactions with cDNAs and long oligonucleotides. Use HybIt® to produce superior data with microarrays printed on atomically flat Super Microarray Glass Substrate Slides.
Table of Contents
Introduction
Quality Control
Product Description
Technical Assistance
Short Protocol
Complete Protocol
Literature Cited
Requirements
Troubleshooting Tips
Ordering Information
Warranty
Introduction
Congratulations on taking a big step towards improving the economies of scale, quality and speed of your genomics research. This booklet contains a complete set of protocols outlining the steps and principles needed to use Arrayit HybIt® Hybridization Solution.
Quality Control
Arrayit assures the performance of this product. The finest scientific research went into the development of this product. Rigorous quality control monitoring on a lot-by-lot basis guarantees that the ingredients conform to the highest industry standards.
Product Description
The Arrayit HybIt® Hybridization Solution is an advanced buffering system containing a patent-pending mixture of salts, detergents, accelerants, and stabilizing agents. HybIt® Hybridization Solution increases the efficiency and specificity of microarray hybridization reactions by expediting base pair formation between complementary target sequences attached to the microarray surface and labeled probe molecules in solution. Users will appreciate the following features:
- Increases hybridization signals by accelerating reaction kinetics.
- Optimized for cDNAs and oligonucleotides.
- Increases detectivity (sensitivity) by reducing background fluorescence.
- Reduces surface tension enabling a uniform hybridization layer.
- Buffering components stabilize extended hybridizations.
- Compatible with many different surface chemistries.
- Arrives as a pre-mixed 1.25X solution that is sterile and ready to use.
- Costs <$1 dollar per hybridization reaction.
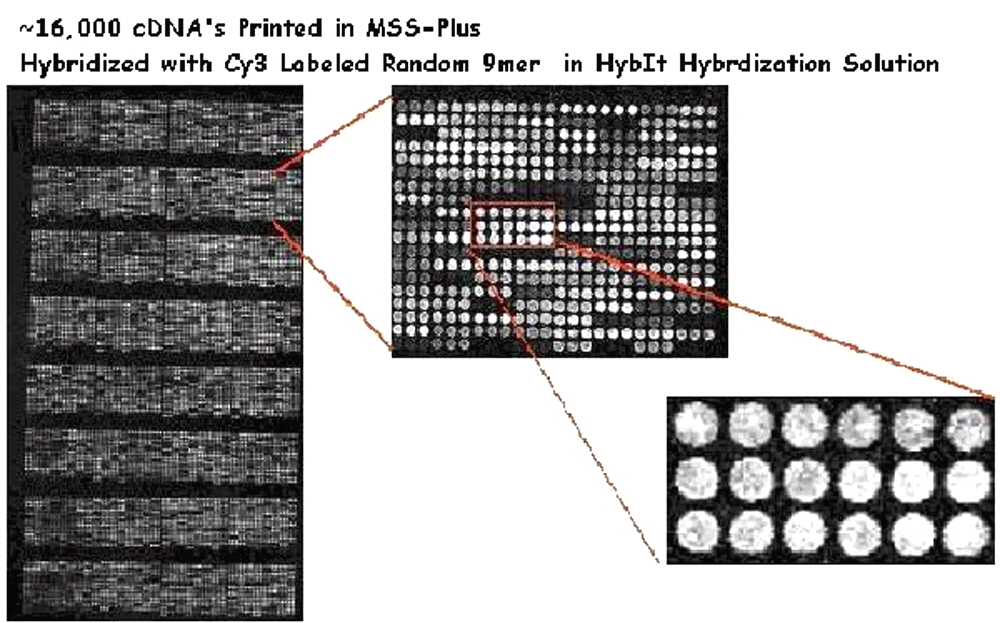
Figure 1. Oligonucleotide hybridization performed using HybIt® Hybridization Solution. A total of 8,000 human cDNAs were purified with Arrayit’s PCR Purification Kit, dried to completion and resuspended in 4.0 µl dH20. A total of 4.0 µl of 2X MSS Plus was added to each sample and the samples were mixed thoroughly by pipetting. The samples were printed on SuperAmine Microarray Substrates, the printed substrates were processed according to the product instructions, and hybridized with a Cy3-labelled random 9-mer at 10 µM concentration in 1X HybIt® Hybridization Solution for 4 hours at room temperature (22°C). The microarray was washed and scanned for fluorescence emission. The quality of the fluorescent signals are easily observed.
Technical Assistance
Please contact us if you have any comments, suggestions, or if you need technical assistance. By electronic mail: arrayit@arrayit.com (under the subject heading, please type Arrayit technical assistance). By email: arrayit@arrayit.com, Monday–Friday PST 9:00 AM – 6:00 PM. Please remember that we want to hear about your successes!
Short Protocol (Steps 1-8)
1. Obtain a nucleic acid microarray.
2. Process the microarray to prepare for the hybridization reaction.
3. Purify the hybrization probe using a suitable purification method.
4. Resuspend the probe in 1.0 µl dH20 and add 4.0 µl 1.25X HybIt® Hybridization Solution.
5. Hybridize the probe to the microarray.
6. Wash away the unbound probe (Arrayit Wash Buffers recommended).
7. Scan or image the microarray to acquire fluorescent signals.
8. Quantify the hybridization results and model the data.
Complete Protocol (Steps 1-8):
1. Obtain nucleic acid microarrays, either by manufacturing them or purchasing pre-made or custom microarrays. Microarrays can be printed using a suitable motion control technology equipped with Arrayit’s Stealth™ or ChipMaker™ Micro Spotting Technology or an equivalent device. Microarrayers can be purchased from a variety of sources including Virtek Vision (Ontario, Canada), Cartesian Technologies (Irvine, CA), Packard Biochip Technologies (Billerica, MA), GeneMachines (Belmont, CA), and many other providers. HybIt® is formulated to improve hybridization results with microarrays printed on a wide range of different microarray manufacturing systems.
2. Process the microarrays to prepare for the hybridization reaction. When using SuperAldehyde Substrates, make certain to allow the printed substrates to dry overnight before processing. Drying can be carried out on the platen of the microarrayer, provided the humidity is <40%. Printed substrates can also be dried in slide boxes, by keeping the lid slightly ajar overnight. Drying is required for Schiff’s base formation between the amino-lniked DNA to the SuperAldehyde surface. Microarrays printed on SuperAmine Substrates can be used within 1 hour after printing, though the substrates must be baked at 80oC for 80 minutes or crosslinked with ultraviolet light (e.g. Stratagene Stratalinker) to strengthen the attachment of the DNA to the SuperAmine surface. After proper drying and/or baking or U.V. crosslinking, the substrates should be processed to remove unbound DNA. Many different protocols have been used successfully for substrate processing. The SuperAldehyde and SuperAmine Substrate product documentation provides additional information on substrate processing.
3. Prior to hybridization, make certain to purify both fluorescent and non-fluorescent probes to remove contaminants that can lead to increased background. Probes can be purified with the ArrayIt Fluorescent Probe Purification Kit or an equivalent purification system.
4. The purified and dessicated probes should be resuspend in 1.0 µl dH20 and 4.0 µl of 1.25X HybIt® Hybridization Solution. Prior to use, make certain to pre-warm the HybIt® Hybridization Solution for 1 min at 65°C and mix the Solution by inverting the tube several times to make sure the components are mixed properly. For a 20 µl hybridization volume, add 4.0 µl dH20 and 16.0 µl of 1.25X HybIt® Hybridization Solution, and scale the volumes accordingly for larger and smaller probe volumes. Probe molecules can be a variety of fluorescent and non-fluorescent species including single-stranded DNA, RNA and oligonucleotides.
5. Hybridize the probe to the microarray. Hybridization reactions can be performed using glass cover slips and 1.25 µl of probe solution per 1.0 cm2 of cover slip. For best results, add the probe to one edge of the microarray surface, and gently lower the cover slip onto the microarray with fine forceps allowing the probe to sheet evenly across the surface between the cover slip and the microarray substrate. Pre-heating the probe solution to the hybridization temperature (e.g. 65°C) has been shown to reduce background fluorescence. Once the cover slip is lowered onto the microarray, transfer the substrate with cover slip to a pre-warmed Hybridization Cassette containing 10.0-µl dH20 to maintain 100% humidity during the hybridization reaction. A volume of 10 -20 ul dH20 placed under the substrate can also be used to further reduce sample dessication. Seal the cassette and hybridize for 1-12 hrs at the proper temperature. For short oligonucleotides (e.g. 9-mers) a hybridization termperature of 22°C works well and for 15-mers, 42°C yields nice results. Long oligonucleotides and cDNAs are hybridized commonly at 65°C.
6. Wash the hybridized substrate to remove the unbound probe molecules. Remove the microarray from the Hybridization Cassette and transfer the substrate immediately to an Arrayit High-Throughput Wash Station and wash the substrate with the appropriate buffers. Arrayit Wash Buffers A, B and C are recommended for cDNA hybridizations and Arrayit Wash Buffers 1, 2 and 3 are recommended for oligonucleotide hybridizations. “Home-made” buffers will also work well for many applications. For indirect labeling procedures that require “staining steps”, microarrays should be stained prior to moving on to the detection step (Step 7).
7. Once the microarray is washed, it should be scanned or imaged to acquire fluorescent signals. Insert the substrate into the ScanArray™ 5000 (Packard BioChip Technologies). Compatible detection systems are also available from Virtek Vision (Ontario, Canada), Axon Instruments (Hayward, CA), Applied Precision (Issaquah, WA) and several other vendors. Scan the area of the substrate containing the microarray. The scan area, excitation source, laser power and PMT settings should be adjusted with the ScanArray™ software to provide optimal signals. Laser and PMT settings should be chosen to give maximal unsaturated signal with minimal background fluorescence. Typically, laser and PMT settings of 70% and 60-80% respectively yield good results with the ScanArray™ 5000 for most hybridized microarrays.
8. Save the hybridization results and quantify the data. Upload the scanned image tiff file into the ImaGene software (BioDiscovery) or a suitable quantitation package and examine each feature on the microarray for fluorescence intensity. Additional manipulations and mining programs can be used to generate ratios, scatter plots, clusters and the like.
HybIt® References
- Oligonucleotide Microarray-Based Mutation Detection of the K-ras Gene in Colorectal Cancers with Use of Competitive DNA Hybridization: http://www.clinchem.org/cgi/content/full/50/9/1688
- Library on a slide for bacterial comparative genomics: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=394321
- STEC-EPEC Oligonucleotide Microarray: A New Tool for Typing Genetic Variants of the LEE Pathogenicity Island of Human and Animal Shiga Toxin–Producing Escherichia coli (STEC) and Enteropathogenic E. coli (EPEC) Strains: http://www.clinchem.org/cgi/content/abstract/52/2/192
- Development and Applications of a ß-Catenin Oligonucleotide Microarray: http://clincancerres.aacrjournals.org/cgi/content/full/9/8/2920
Literature Cited
1. Chee, M., Yang, R., Hubbell, E., Berno, A., Huang, X. C., Stern, D., Winkler, J., Lockhart, D. J., Morris, M. S., Fodor, S. P. A. (1996) Accessing genetic information with high-density DNA arrays. Science 274: 610-614.
2. de Saizieu, A., Certa, U., Warrington, J., Gray, C., Keck, W., and Mous, J. (1998) Bacterial transcript imaging by hybridization of total RNA to oligonucleotide arrays. Nature Biotech. 16: 45-48.
3. A comprehensive set of nearly 1,000 microarray citations is available in an electronic format at http://arrayit.com/e-library.
Recommended Equipment and Reagents
Stealth™ or ChipMaker™ Micro Spotting Technology
High Throughput Wash Station
Microarray Wash Station
Fluorescent Probe Purification Kit
Micro Spotting Solution Plus
Micro Spotting Solution
Hybridization Cassettes
SuperAldehyde or SuperAmine Substrates
Troubleshooting Tips
High Background:
- Contaminated cDNAs or oligonucleotides used for printing. For PCR products, use the PCR Purification Kits.
- Used conventional buffers instead of HybIt® Hybridization Solution
- Hybridization temperature too low
- Probes were not purified sufficiently to remove unincorporated fluors. Use the Fluorescent Probe Purification Kit.
Reagents - Microarray Buffers - Hybridization Buffers - UniHyb™ Hybridization Solution for Single Nucleotide Polymorphism SNP Microarrays
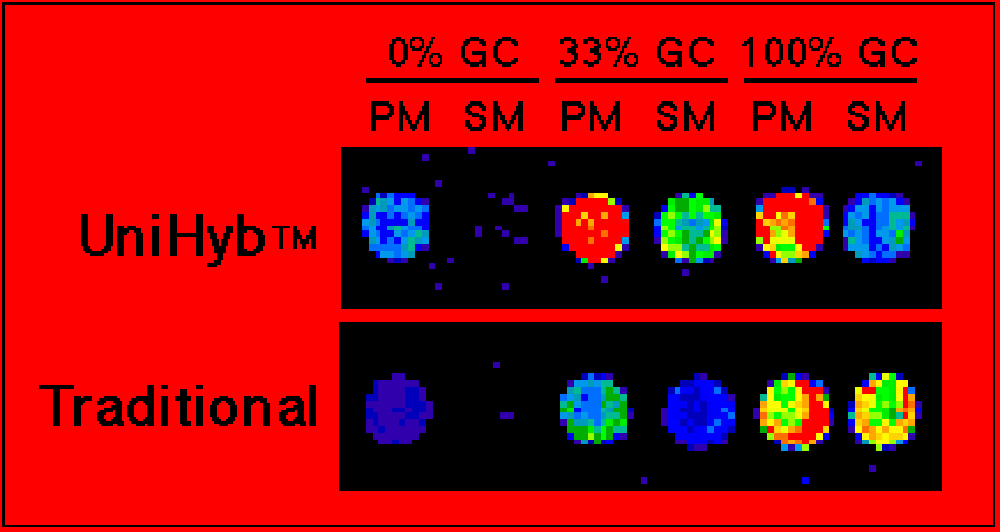
Detect single base mismatches in any sequence! Arrayit has developed UniHyb™ Hybridization Solution for mutation detection, single nucleotide polymorphism (SNP) detection, DNA re-sequencing and sequencing by hybridization. UniHyb™ accelerates the rate of hybridization, reduces background and minimizes energetic differences between G-C and A-T base pairs, making UniHyb™ ideal for all microarray genotyping applications.
Table of Contents
Introduction
Quality Control
Product Description
Technical Assistance
Short Protocol
Complete Protocol
Literature Cited
Requirements
Troubleshooting Tips
Ordering Information
Warranty
Introduction
Congratulations on taking a big step towards improving the economies of scale, quality and speed of your genomics research. This booklet contains a complete set of protocols outlining the steps and principles needed to use Arrayit UniHyb™ Hybridization Solution.
Quality Control
Arrayit assures the performance of this product. The finest scientific research went into the development of this product. Rigorous quality control monitoring on a lot-by-lot basis guarantees that the ingredients conform to the highest industry standards.
Product Description
The Arrayit UniHyb™ Hybridization Solution is an advanced hybridization solution containing a patent-pending mixture of salts, detergents and buffering components. Use of UniHyb™Hybridization Solution will increase the quality of microarray biochip hybridization reactions involving base pairing interactions between complementary nucleic acid chains.
Users will appreciate the following features:
- Supports multiple microarray technologies
- Increases signal by accelerating hybridization kinetics
- Increases sensitivity by reducing background fluorescence
- Minimizes sequence context effects
- Increases hybridization specificity
- Reduces surface tension providing a uniform hybridization layer
- Buffering components stabilize extended reactions
- Compatible with many glass-based surface chemistries
- Arrives pre-mixed and sterile, no preparation required
- Costs <$1 dollar per hybridization reactionFigure 1. Mutation detection. Shown are scanned images of oligonucleotide microarrays printed with Arrayit's ChipMaker™ 3 Micro Spotting device. Spacing is 150 µm center-to-center on silylated slides. The oligonucleotides are three pairs of amino-linked 15-mers bearing a perfect match (PM) or a single base mismatch (SM) relative to a Cy3-labeled probe. Hybridizations were performed for 4 hrs at 42°C with 0.2 pmole/µl probe in Unihyb™ Hybridization Solution (top) or 5X SSC + 0.2% SDS (bottom). Fluorescent detection was performed using the ScanArray 3000 from General Scanning, Inc. Improvements in hybridization signal and specificity are easily observed with Unihyb™ Hybridization Solution compared to a conventional hybridization buffer. Point mutations are easily identified in sequences spanning the entire range of AT and GC base composition.
Technical Assistance
Please contact us if you have any comments, suggestions, or if you need technical assistance. By electronic mail: arrayit@arrayit.com (under the subject heading, please type “Arrayit technical assistance”). By telephone: (408) 977-0160, Monday–Friday PST 9:00am - 4:30pm. Please remember that we want to hear about your successes!
Short Protocol (Steps 1-7)
1. Print oligonucleotide microarrays.
2. Process the oligonucleotide microarrays for hybridization.
3. Resuspend fluorescent probe in 1.0 part dH20 and 4.0 parts UniHyb™ Solution.
4. Hybridize the probe to the microarray under the appropriate conditions.
5. Wash away the unbound fluorescent probe.
6. Scan the microarray for fluorescent signal.
7. Score the hybridization results.
Complete Protocol (Steps 1-7)
1. Print oligonucleotide microarrays with the ChipMaker™, Stealth, 946 or Professional Micro Spotting device or a suitable microarray technology. In principle, UniHyb™ should improve mutation detection on many different microarray platforms. For mutation detection with spotted arrays, 15-mers provide ideal discrimination, though oligonucleotides of 7 to 25 nucleotides will also work. Oligonucleotide 15-mers bearing an amino-linker on the 5’ end will attach readily to silylated slides. A 10 pmole/µl oligonucleotide in 1X Arrayit Micro Spotting Solution will provide high quality microarrays suitable for mutation detection. Mismatches at the center position (8th nucleotide) of a 15-mer provide the highest degree of discrimination, though mismatches throughout the oligonucleotide will also allow mutation detection to a lesser degree.
2. Process the oligonucleotide microarrays for hybridization. After the oligonucleotides are spotted, the slides should be allowed to dry at room temperature overnight. This can be accomplished by placing the slides in a slide box with the lid slightly ajar. Drying increases the binding of the amino-modified oligonucleotide to the SuperAldehyde or SuperEpoxy surface. After drying, the slides should be processed to remove unbound oligonucleotide. Many protocols have been used for slide processing, though the one given below works well and is easy to implement. Transfer six slides to a Arrayit Wash Station and place the Wash Station and slides in a 600 ml beaker with a stir bar containing 500 ml of each buffer. Wash twice for 2 min each at room temperature in 0.2% SDS, twice for 2 min each at room temperature in dH20, once for 2 min at 95-100°C in dH20, cool to room temperature for 1 min, once for 5 min at room temperature in sodium borohydride solution (1.3 g NaBH4 dissolved in 375 ml phosphate buffered saline {PBS = Sigma Cell Culture Catalog # D8537}, then add 125 ml pure ethanol), three times for 1 min each at room temperature in 0.2% SDS, twice for 1 min each at room temperature in dH20. Air dry the slides to completion. Slides are ready for hybridization.
3. Resuspend the fluorescent probe in 1X UniHyb™ Hybridization Solution, which is provided as a 1.25X solution. This is accomplished by first resuspending the probe in 1.0 part dH20, then adding 4.0 parts of UniHyb™ Hybridization Solution. Do not attempt to re-suspend the probe in neat UniHyb™. Prior to using UniHyb™ Hybridization Solution, pre-warm the solution for 30 sec at 65°C and mix by inverting the tube several times to re-dissolve the detergents present in the UniHyb™. Failure to pre-warm the UniHyb™ prior to use may lead to poor hybridization results! A fluorescent probe desiccated to dryness would be re-suspended by adding 2.0 µl of dH20, followed by 8.0 µl of pre-warmed 1.25X UniHyb™ Hybridization Solution. Probes should be single-stranded DNA or RNA molecules made by either PCR or in vitro transcription of RNA. Probes containing polynucleotides 15-50 nt in length give superior hybridization results for mutation detection experiments compared to longer polynucleotides.
4. Hybridize the probe to the microarray under the appropriate conditions. This is accomplished by using 1.25 µl of probe in 1X UniHyb™ per cm2 glass cover slip. For best results, add the probe to one edge of the cover slip surface, then gently lower the cover slip onto the microarray with fine forceps allowing the probe to sheet evenly across the surface between the cover slip and the slide. Transfer the slide with cover slip to an Arrayit Hybridization Cassette containing 3.0 µl dH20, seal the cassette and hybridize for 0.5-4.0 hrs at the appropriate temperature. The hybridization temperature should be ~10°C below the Tm of the average heteroduplex on the microarray. Arrayit recommends the following hybridization temperatures with UniHyb™ Hybridization Solution: 15-mers (42°C), 17-mers (47°C), 19-mers (52°C), 21-mers (55°C), 23-mers (57°C), 25-mers (60°C). Shorter or longer oligonucleotides will utilize lower or higher temperatures, respectively.
5. Wash away the unbound fluorescent probe. Remove the microarray from the Hybridization Cassette and immediately transfer the slide to an ArrayIt Wash Station positioned in a 600 ml beaker containing 500 ml of 2X SSC + 0.2% SDS with constant mixing. After 1 min of mixing, gently remove the cover slip from the surface of the slide using a fine forceps. This can be accomplished by applying gentle pressure to the surface of the cover slip with the forceps, and sliding the cover slip off the surface of the microarray. Do not allow the forceps to contact the surface of the microarray directly. Direct contact can lead to scratches and poor data! Wash the slides once for 15 min at room temperature in 2X SSC + 0.2% SDS, then once for 5 min at room temperature in 2X SSC. Allow the slides to air dry for 15 min and blot off any remaining liquid.
6. Scan the microarray for fluorescent signals. Insert the 1" X 3" slide into the ScanArray 3000 (GSI Lumonics) or a compatible detection system and scan the area of the slide containing the microarray. The scan area, excitation source, laser power and PMT settings can all be adjusted with the ScanArray software. Laser and PMT settings should be chosen to give maximal unsaturated signal with minimal background fluorescence. Typically, laser and PMT settings of 70% and 60-80% respectively yield good results with the ScanArray 3000.
7. Score the hybridization results. Upload the scanned image tiff file into the ImaGene software (BioDiscovery) or a suitable quantitation package and examine each feature for fluorescence intensity. Oligonucleotides that form a perfect match (PM) with the fluorescent probe will produce greater fluorescence intensity than any of the single base mismatches (SM) oligonucleotides. The signal of the perfect match to a single base mismatch (SM) is 50:1-10:1 depending on the sequence.
UniHyb References
- Genes transactivated by hepatitis C virus core protein, a microarray assay -
M Liu, SL Zhang, J Cheng, Y Liu, L Wang, Q Shao, J … - World J Gastroenterol, 2005 - wjgnet.com... Hybridization conditions Hybridization of the fluorescent probe to the microchipwas performed in 1× UniHyb solution at 37 for 30 min. ... - Microarray Analysis of Microbial Virulence Factors -
V Chizhikov, A Rasooly, K Chumakov, DD Levy - Applied and Environmental Microbiology - aem.asm.org... Hybridization conditions. Hybridization of the fluorescent probe to the microchip was performed in 1× UniHyb solution (ArrayIt) at 37°C for 30 min. - …Molecular Detection and Identification of Influenza Viruses by Oligonucleotide Microarray - S Sengupta, K Onodera, A Lai, U Melcher - Journal of Clinical Microbiology - jcm.asm.org... in 2 µl of water, and denatured for 5 min at 100°C. It was snap-cooled on ice and mixed with 8 µl of preheated (65°C for 3 min) Unihyb hybridization buffer ...
- …RET Oligonucleotide Microarray for the Detection of RET Mutations in Multiple Endocrine Neoplasia - IJ Kim, HC Kang, JH Park, JL Ku, JS Lee, HJ Kwon, … - Clinical Cancer Research, 2002 - clincancerres.aacrjournals.org... Prepared PCR products of each exon were mixed, resuspended in prewarmed 1 x UniHyb Solution (Arrayit International Inc., Sunnyvale, CA) at 3µ , and ...
- …The food entrainable oscillator studied by DNA microarrays: What is the liver doing during food - AF Báez-ruiz, DF Luna-Moreno, OF Vázquez- … - Biological Rhythm Research, 2005 - Taylor & Francis... Equal quantities of labeled cDNA were hybridized, using hybridization solution UniHyb(Arrayit International Inc.), to the collection of ‘‘Rat Liver Arrays ...
- Analysis of differentially expressed transcripts from planthopper-infested wild rice (Oryza minuta) -
SKJ Cho, KWJ Jung, JUJ Jeung, KHJ Kang, KSJ Shim, … - Plant Cell Reports, 2005 - Springer... The pu- rified labeling mixture was diluted with 1.25”? ArrayIt UniHyb solution (Arrayit, Sunnyvale, Calif.; http://www.arrayit.com) and hybridized at 55 C ... - Detection and Identification of Mycobacterium Species Isolates by DNA Microarray - »group of 4
M Fukushima, K Kakinuma, H Hayashi, H Nagai, K Ito … - Journal of Clinical Microbiology - jcm.asm.org
... The fluorescently labeled RNA was resuspended in 2.0 µl of sterile water and then
in 8.0 µl of prewarmed 1.25x UniHyb hybridization solution (Arrayit ...
Cited by 23 - Web Search - BL Direct - …Mutational analysis of OGG1, MYH, MTH1 in FAP, HNPCC and sporadic colorectal cancer patients: R154H - »group of 4
IJG Kim, JLG Ku, HCG Kang, JHG Park, KAG Yoon, YG … - Human Genetics, 2004 - Springer
... Amplified products were re-suspended in 3.5 μl pre-warmed 1.25×UniHyb Solution
(Arrayit International, Sunnyvale, Calif.), and hybridized in a saturated ...
Cited by 1 - Web Search - …Arrayed Primer Extension: A Robust and Reliable Genotyping Platform for the Diagnosis of Single Gene - »group of 4
YI LU, SKOWYIN KHAM, P TAN, TC QUAH, CK HENG, … - GENETIC TESTING, 2005 - liebertonline.com
... of single-stranded DNA amplified from each of the -globin gene and of TPMT gene
fragments were mixed and diluted in 8 l UniHyb™ Hybridization Solution ...
Web Search - …Detection and identification of intestinal pathogenic bacteria by hybridization to oligonucleotide - LQ Jin, JW Li, SQ Wang, FH Chao, XW Wang, ZQ Yuan - World J Gastroenterol, 2005 - wjgnet.com... microarrays using the following protocol: 1 μL of amplicons was added into a tube containing 4 μL hybridization solution (UniHyb TM , Arrayit International ...
- GENE EXPRESSION PROFILING BY DNA MICROARRAY TECHNOLOGY - »group of 2
K Iida, I Nishimura - Critical Reviews in Oral Biology & Medicine - crobm.iadrjournals.org... Sup and Printheads, Hybridization Cassette, and ChipMaker Pins uniHyb Hybridiation Sub Arrayit International, Inc. and Printheads Solution, Wash Solution ... - Metagenomic Profiling: Microarray Analysis of an Environmental Genomic Library -
JL Sebat, FS Colwell, RL Crawford - Applied and Environmental Microbiology - aem.asm.org... g of salmon sperm genomic DNA and reduced to a volume of 5 µl by using Microcon
YM-30 concentrators (Millipore); 20 µl of 1.25x unihyb hybridization buffer ... - The whole EST catalog - B Klevecz - The Scientist, 1999 - the-scientist.com
... Arrayit also offers proprietary microspotting and hybridization buffers, including Unihyb, which expedites mismatch detection with microarrays. ... - Molecular Biology for the Environment: an EC-US hands-on Course in Environmental Biotechnology
V de Lorenzo, JL Ramos, J Kukor, GJ Zylstra - Conference: Molecular Biology for the Environment: an EC-US …, 2004 - osti.gov Page 1. MOLECULAR BIOLOGY FOR THE ENVIRONMENT A short EC-US Course in Environmental Biotechnology Under the auspices of the EC-US ... - DO-IT-YOURSELF CD Advanced laboratory and bioinformatics methods in environmental biotechnology
F Agencies, F de Ciencias, C de Cantoblanco, S … - osti.gov
Page 1. MOLECULAR BIOLOGY FOR THE ENVIRONMENT A short EC-US Course in
Environmental Biotechnology Under the auspices of the EC-US ... - …DEVELOPMENT OF MICROSYSTEM TECHNOLOGY SUITABLE FOR BACTERIAL IDENTIFICATION AND GENE EXPRESSION
DESAG ZUR ERLANGUNG - deposit.ddb.de
Page 1. DEVELOPMENT OF MICROSYSTEM TECHNOLOGY SUITABLE FOR BACTERIAL
IDENTIFICATION AND GENE EXPRESSION MONITORING DISSERTATION ZUR
Literature Cited
Chee, M., Yang, R., Hubbell, E., Berno, A., Huang, X. C., Stern, D., Winkler, J., Lockhart, D. J., Morris, M. S., Fodor, S. P. A. (1996) Accessing genetic information with high-density DNA arrays. Science 274: 610-614.
de Saizieu, A., Certa, U., Warrington, J., Gray, C., Keck, W., and Mous, J. (1998) Bacterial transcript imaging by hybridization of total RNA to oligonucleotide arrays. Nature Biotech. 16: 45-48.
Drmanac, S., Kita, D., Labat, I., Hauser, B., Schmidt, C., Burczak, J.D., Drmanac, R. (1998) Accurate sequencing by hybridization for DNA diagnostics and individual genomics. Nature Biotech. 16, 54-58.
Hacia, J. G., Brody, L. C., Chee, M. S., Fodor, S. P. A., Collins, F. S. (1996) Detection of heterozygous mutations in BRCA1 using high density oligonucleotide arrays and two-colour fluorescence analysis. Nature Genet. 14: 441-447.
Khrapko, K.R., Khorlin, A.A., Ivanov, I.B., Chernov, B.K., Lysov, Yu.P., Vasilenko, S.K., Florent’ev, V.L., Mirzabekov, A.D. (1991) Hybridization of DNA with oligonucleotides immobilized in gel: a convenient method for detecting single base substitutions. Molecular Biology 25: 581-591.
Maskos, U., Southern, E.M. (1992) Oligonucleotide hybridizations on glass supports: a novel linker for oligonucleotide synthesis and hybridization properties of oligonucleotides synthesised in situ. Nucleic Acids Res. 20: 1679-1684.
Vladimir Chizhikov, Avraham Rasooly, Konstantin Chumakov, and Dan D. Levy Microarray Analysis of Microbial Virulence Factors Appl. Envir. Microbiol. 2001 67: 3258-3263.
Recommended Equipment and Reagents
Chipmaker or Stealth Micro Spotting Dev
HT Wash Station
Micro Spotting Solution
Hybridization Cassettes
Microarray SuperAldehyde Substrates
Troubleshooting Tips
Poor mutation detection:
o Impure oligonucleotides used for spotting
o Used conventional buffers instead of UniHyb Hybridization Solution
o Incorrect hybridization temperature

