VIP™ Microarrays
Data Sheet
![]() Shop this product in our online store
Shop this product in our online store
Arrayit VIP™ technology allows the identification of any amplified nucleic acid sequence from any organism. The main clinical application areas include population screening, disease screening, disease diagnosis, parentage testing, human leukocyte antigen (HLA) typing, blood typing, infectious disease diagnosis, forensics, and neonatal and adult screening. VIP™ technology can also be applied to a myriad of research areas including genetic studies in human, mouse, rat, higher plants, bacteria and viruses aimed at detecting specific sequences, or minor sequence variants including single nucleotide polymorphisms (SNPs), mutations, insertions, deletions and insertion-deletion (indel) mutations. VIP™
Technology Overview
Arrayit VIP™ technology is a universal microarray analysis platform for nucleic acid-based genetic screening, testing, diagnostics, genotyping and single nucleotide polymorphism (SNP) analysis. Arrayit technology allows hybridization-based analysis of SNPs, mutations, deletions, insertion-deletion mutations (indels), and other minor sequence variants from any organism of interest. Arrayit also allows detection of bacterial and viral sequences as required for the diagnosis of infectious diseases caused by agents such as mycobacterium tuberculosis, cytomegalovirus (CMV), human immunodeficiency virus (HIV), bacillus anthracis, and other pathogens.
Specific genetic loci are amplified by use of the polymerase chain reaction (PCR), printed into a microarray, and hybridized with fluorescent synthetic oligonucleotides. The hybridized microarrays are then scanned for fluorescence emission to derive genotyping information (Figure 1). Normal, carrier and disease genotypes are distinguished easily because slight changes in primary DNA sequence alter hybridization efficiency (Figure 2). Altered hybridization efficiency leads to different fluorescent intensities in the scanned microarray image (Figure 3). Multi-color strategies involving cyanine3 and cyanine5 fluorescent labels can be used to improve the readability and precision of the assays (Figure 3). If performed correctly, the Arrayit method is 100% accurate. A microarray containing 25,000 individual printed features showcases the extraordinary quality of Arrayit® microarrays (Figure 4). Platinum, Gold, Silver and Bronze Cleanroom plans provide integrated environments designed to meet the most exacting clinical and research applications. Visit the Arrayit Diagnostics Division for Platform details.
The Arrayit approach is clear departure from traditional microarray assays that use oligonucleotide capture probes to analyze one or two patient samples at a time. Instead of the patient samples floating in solution, amplified material from each patient is attached at a distinct location to the microarray substrate, allowing massively parallel analysis of multiple patients and multiple loci in a single test. The current printing capacity (>100,000 spots per microarray) enables screening of 10,000 patients at 10 different loci on one chip. Arrayit is the only platform that allows the economical screening of millions of patients per year. This revolutionary "samples down" approach allows all of the costs of the test (hardware, software, labor, kits, reagents, etc.) to be amortized across many patients to provide a cost-effectiveness that is impossible to achieve with traditional methods. Arrayit technology can be scaled down to allow testing of tens or hundreds of samples per chip, rendering it ideal for hospitals and public health facilities that process a relatively small number of samples, but require testing on a daily basis. Arrayit technology is rapid, providing accurate genotyping information within 24-48 hours of receiving a patient sample. In the future, Arrayit is expected to integrate into the larger healthcare picture, enabling physicians and other health care officials to make increasingly better clinical decisions.
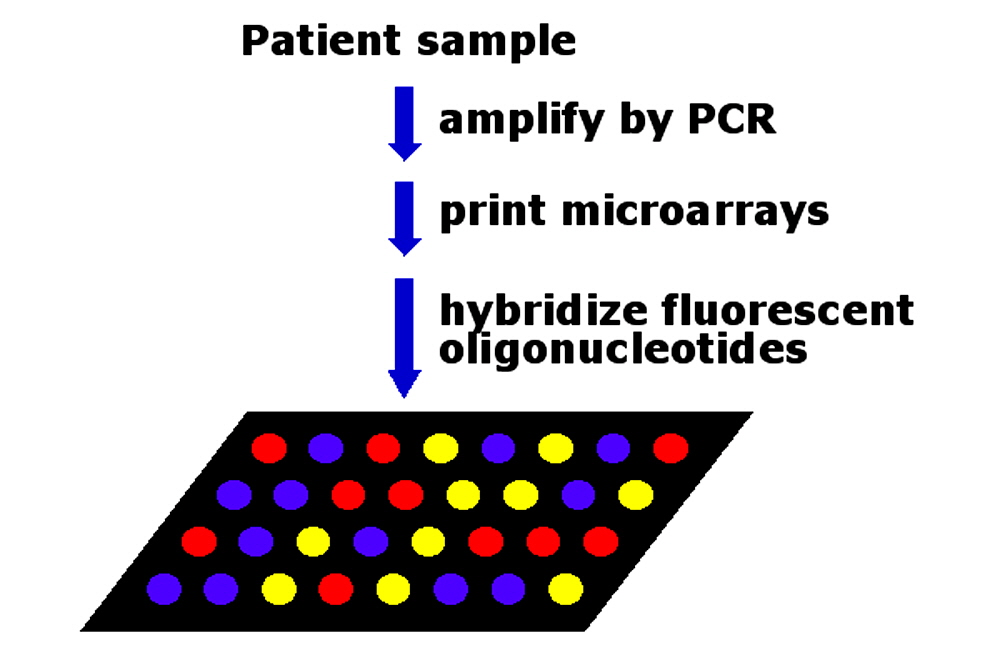
Figure 1. VIP™ Method. The patented Arrayit method is a universal microarray-based platform for genetic screening. Specific chromosomal loci are amplified by use of the polymerase chain reaction (PCR), printed into microarrays, and hybridized with fluorescent oligonucleotides. The fluorescent microarrays are then scanned for fluorescence emission and signal strengths provide genotyping information.

Figure 2. VIP™ is specific for single nucleotide changes. Amplified DNA segments (black letters) from patients with normal (G/G), carrier (G/A) and disease (A/A) genotypes are identical except at a single nucleotide position, which is G in the normal allele and A (red letter) in the mutated allele. Complementary fluorescent oligonucleotides (green letters) hybridized with different efficiencies to the three samples, producing strong, intermediate and weak signals, respectively, for the normal, carrier and disease patients.
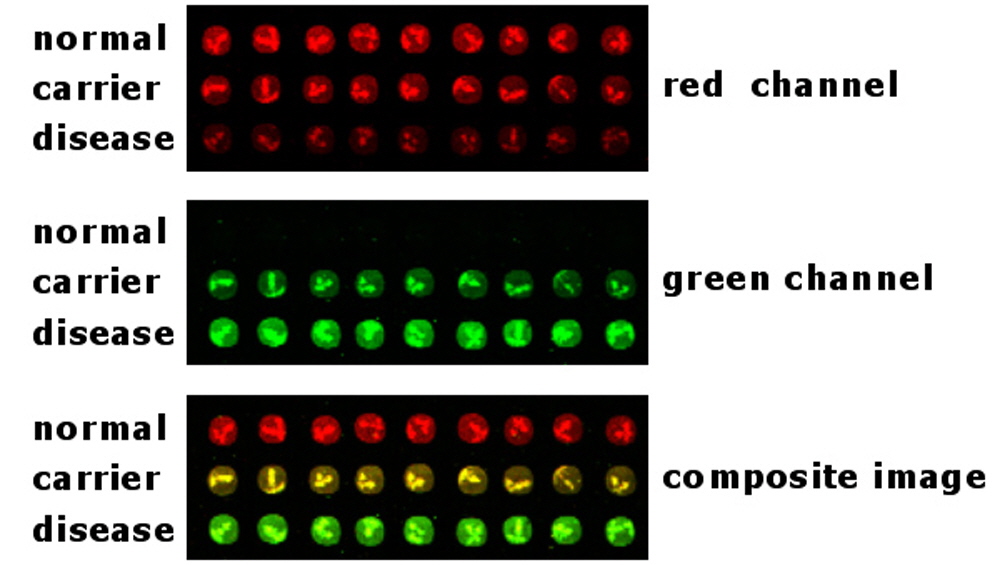
Figure 3. VIP™ 2-color strategies. The VIP™ method was used to analyze samples from patients with different disease genotypes. Fluorescent oligonucleotides bearing cyanine5 (red channel) and cyanine3 (green channel) were co-hybridized to a microarray, scanned for cyanine3 and cyanine5 emission, and the fluorescent images were superimposed to produce a composite, 2-color image (bottom panel). The 3 different genotypes are distinguished easily by scoring the red, yellow and green colors.
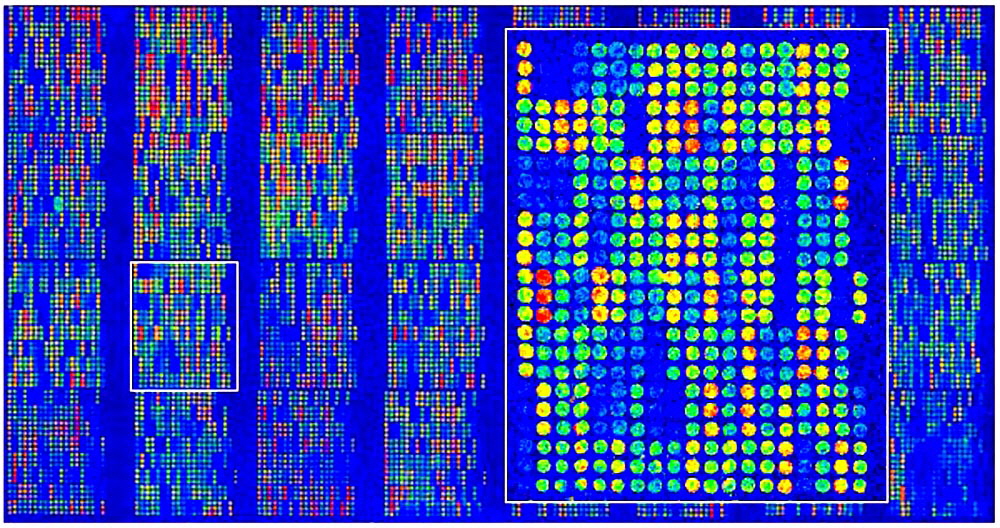
Figure 4. VIP™ microarray with 25,000 features. Arrayit Diagnostics manufacturing and cleanroom technology enable printing of extremely high-quality microarrays, which improves the precision and reliability of the data. Chips containing 25,000 individual features (shown here) allow the simultaneous genotyping of 25,000 different patients at one locus in a single test.

